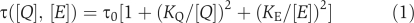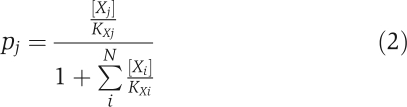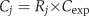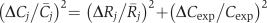Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli (original) (raw)
Abstract
Despite extensive study of individual enzymes and their organization into pathways, the means by which enzyme networks control metabolite concentrations and fluxes in cells remains incompletely understood. Here, we examine the integrated regulation of central nitrogen metabolism in Escherichia coli through metabolomics and ordinary-differential-equation-based modeling. Metabolome changes triggered by modulating extracellular ammonium centered around two key intermediates in nitrogen assimilation, α-ketoglutarate and glutamine. Many other compounds retained concentration homeostasis, indicating isolation of concentration changes within a subset of the metabolome closely linked to the nutrient perturbation. In contrast to the view that saturated enzymes are insensitive to substrate concentration, competition for the active sites of saturated enzymes was found to be a key determinant of enzyme fluxes. Combined with covalent modification reactions controlling glutamine synthetase activity, such active-site competition was sufficient to explain and predict the complex dynamic response patterns of central nitrogen metabolites.
Keywords: active-site competition, flux regulation, HPLC-MS, metabolic dynamics, predictive model
Introduction
Microbes tailor their growth rate to the nutrient environment. For example, Escherichia coli grow faster in the presence of ample ammonium than when nitrogen is scarce (Atkinson et al, 2002; Gyaneshwar et al, 2005). Although the metabolic pathways converting nutrients to biomass are well known, a quantitative understanding of the relationship between nutrient environment, metabolism, and growth rate is still missing. Furthermore, despite much progress towards quantitatively understanding potential mechanisms for controlling metabolite concentrations and fluxes (as reviewed in Sorribas and Savageau, 1989; Heinrich and Schuster, 1996; Fell, 1997), the most important control mechanisms operating in cells have not been rigorously dissected. This is particularly true for networks containing cycles and branches.
One approach to gaining quantitative understanding of cellular metabolic regulation is to analyze the dynamics of intracellular metabolite concentrations in response to changes in nutrient availability. For such analysis, nitrogen assimilation in E. coli provides a tractable model network. As a prokaryote, E. coli lacks extensive compartmentation. Compared with carbon metabolism, the metabolic circuitry for nitrogen assimilation is simpler. Extensive biochemical analysis has been conducted on some of the key nitrogen assimilation enzymes in E. coli, especially glutamine synthetase (GS) (Miller and Stadtman, 1972; Schutt and Holzer, 1972; Mantsala and Zalkin, 1976; Kustu et al, 1984; Alibhai and Villafranca, 1994). A strong correlation has been demonstrated between the intracellular pool size of glutamine and growth rate under nitrogen limitation in the closely related organism Salmonella (Ikeda et al, 1996).
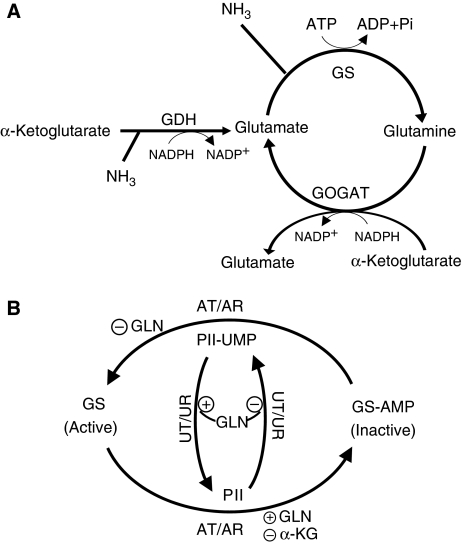
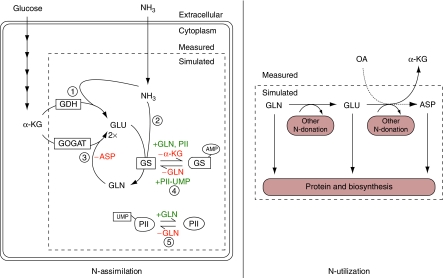
The networks of ammonium assimilation and GS regulation in E. coli are shown in Figure 1. Glutamate is the principal cellular nitrogen distributor (Ikeda et al, 1996; Reitzer, 2003) and can be produced through either a single reaction catalyzed by glutamate dehydrogenase (GDH) or by the glutamine synthetase/glutamate synthase cycle (GS/GOGAT) (Figure 1A). GDH directly converts α-ketoglutarate, ammonia, and reducing power (NADPH) to glutamate. The GS/GOGAT cycle catalyzes the same net reaction, but by first producing glutamine, and then converting one glutamine and one α-ketoglutarate to two molecules of glutamate. The GDH reaction does not consume ATP, whereas the GS reaction does. In exchange for this energy input, GS has a higher affinity than GDH for ammonia (_K_m∼0.1 versus 1.5 mM) (Sakamoto et al, 1975; Alibhai and Villafranca, 1994).
Figure 1.
Ammonia assimilating pathways and regulation of glutamine synthetase in E. coli. (A) Ammonia can be assimilated through either glutamate dehydrogenase (GDH) or the glutamine synthetase/glutamate synthase cycle (GS/GOGAT). For each turn of the GS/GOGAT cycle, one molecule of ammonia is assimilated into glutamate. (B) GS catalytic activity is regulated by the bifunctional enzyme adenylyltransferase/adenylyl-removing enzyme (AT/AR), with adenylylation inactivating GS. The activity of AT/AR is regulated by the signaling protein PII, which is itself covalently modified by another bifunctional enzyme, uridylyltransferase/uridylyl-removing enzyme (UT/UR). Unmodified PII indicates nitrogen sufficiency and promotes GS adenylylation. Glutamine and α-ketoglutarate allosterically regulate AT/AR and UT/UR as shown.
GS is extensively regulated. Its transcription and catalytic activity are controlled by two interlinked cycles of protein covalent modification that respond to the intracellular concentrations of glutamine and α-ketoglutarate (Figure 1B) (Schutt and Holzer, 1972; Senior, 1975; Garcia and Rhee, 1983; Kustu et al, 1984; Atkinson et al, 2002; Reitzer, 2003; Jiang et al, 2007). The ultimate function of these cycles is to regulate the concentration of active (unmodified) GS. On short time scales, this is largely achieved through GS adenylylation/deadenylylation by the bifunctional adenylyltransferase/adenylyl-removing enzyme (AT/AR) (Schutt and Holzer, 1972; Kustu et al, 1984; Reitzer, 2003). AT/AR is controlled directly by glutamine (which favors adenylylation, or inactivation), and also by the signaling protein PII (which in its unmodified state indicates nitrogen sufficiency, favoring GS adenylylation through AT/AR and also turning off GS transcription) (Bancroft et al, 1978; Reitzer, 2003; Ninfa and Jiang, 2005). PII can itself be covalently modified (thereby indicating nitrogen limitation) by another bifunctional enzyme, uridylyltransferase/uridylyl-removing enzyme (UT/UR), which uridylylates PII when glutamine is scarce (Jiang et al, 1998a; Ninfa and Atkinson, 2000). In addition, PII can be modified by noncovalent binding of α-ketoglutarate, which rises in concentration during nitrogen limitation (Ninfa and Jiang, 2005; Brauer et al, 2006). PII can also respond to other inputs, including adenylylate energy charge (Jiang and Ninfa, 2007).
The net effect of the PII-AT/AR-GS cascade on nitrogen assimilation (on short time scales, in which transcriptional regulation is not important) has been simulated computationally by Bruggeman et al, who developed an ordinary-differential-equation model of the network (Bruggeman et al, 2005). Kinetic parameters of the model were estimated based on biochemical studies of individual components, conducted in large part by Ninfa and colleagues (Jiang et al, 1998a, 1998b; Ninfa and Atkinson, 2000). This modeling effort revealed that the system is indeed capable of producing the anticipated regulatory behavior: under nitrogen poor conditions, PII is uridylylated and GS is not covalently modified, whereas under conditions of nitrogen excess, PII is unmodified and GS is substantially inactivated by adenylylation. Moreover, the model showed that such regulation is adequate to control glutamine and glutamate concentrations, with glutamine varying more widely in response to nitrogen availability than glutamate, consistent with experimental data (Schutt and Holzer, 1972; Ikeda et al, 1996).
Although informative, the model of Bruggeman et al is incomplete in several respects. It considers the concentration of α-ketoglutarate to be constant during nitrogen perturbations. As we show here, the α-ketoglutarate pool size rapidly changes by an order-of-magnitude in response to changing nitrogen availability. Given that α-ketoglutarate is both a substrate for nitrogen assimilation and a regulator of the GS covalent modification cascade, considering its dynamics is critical. In addition, Bruggeman et al assume that consumption of glutamine and glutamate are independent of each another, which fails to take into account the stoichiometric requirements for each in building biomass. Thus, Bruggeman et al fail to capture the fundamental interdependence of metabolite pools and growth: metabolite pool sizes can control growth rate, with growth requiring consumption of metabolites in fixed ratios dictated by the composition of biomass. The integrated control of nitrogen assimilation within the larger framework of overall cellular metabolic activity therefore remains an open question.
Here, we investigate the short-term (i.e., nontranscriptional) regulation of ammonium assimilation in E. coli using a combination of experiments and computational modeling. We sample the metabolome of ammonium-limited and ammonium-replete E. coli and use mass spectrometry to quantify a broad spectrum of cellular metabolites in the resulting extracts. We then use data on central nitrogen metabolites to drive the development of a dynamic model that links extracellular ammonium availability to intracellular metabolite concentrations and thereby cellular growth rate. Through this data-driven modeling process, we demonstrate that competition for enzyme active sites by substrates, products, and inhibitors is an important component regulating cellular nitrogen assimilation fluxes. Thus, even for one of the best-studied metabolic subnetworks, combining metabolomic experiments with modeling identified an important but previously overlooked means of flux control.
The resulting model quantitatively reproduces the experimentally observed metabolic responses used in model development. In addition, it accurately predicts cellular responses to different perturbations. This work thereby lays the groundwork for combining metabolomics and modeling to develop larger predictive models of metabolic dynamics.
Results
GS/GOGAT cycle produces most of the glutamate in wild-type E. coli grown on sufficient glucose and ammonium
Determination of the pathways that carry the bulk of steady-state flux is a prerequisite to understanding metabolic dynamics. To investigate the relative contribution of the GDH and GS/GOGAT pathways to glutamate synthesis in cells grown with ample glucose and ammonium, we have previously used an isotope tracer-based approach, kinetic flux profiling, which suggested a dominant role for the GS/GOGAT cycle (Yuan et al, 2008). To investigate whether GS/GOGAT indeed predominates, here we conducted kinetic flux profiling of E. coli mutants lacking GDH or GOGAT.
In media with adequate glucose and ammonium, ΔGDH and ΔGOGAT grew comparably to wild type (Supplementary Figure 1A), and also showed a similar overall metabolome when compared with wild type (Supplementary Figure 1B). Consistent with GDH carrying little flux, knocking out GDH did not alter the kinetics of cellular assimilation of 15N-ammonium into glutamine or glutamate (Supplementary Figure 1C and D). In contrast, knocking out GOGAT substantially slowed assimilation of 15N-ammonium into glutamine (Supplementary Figure 1C), with flux through glutamine reduced from 54 mM/min to 13 mM/min (Supplementary Table 1). The results are consistent with 41 mM/min of glutamine feeding into GOGAT in wild type, and generating 82 mM/min of glutamate (because of the 1:2 glutamine:glutamate stoichiometry of GOGAT), which is >85% of the total glutamate biosynthetic flux in wild type (Yuan et al, 2006). Thus, E. coli uses the GS/GOGAT cycle as the major route for glutamate production, even when the more energy-efficient GDH pathway can support equally rapid growth.
Ammonium limitation of filter-grown cultures
We grew E. coli on filters on top of an agarose-media mixture to enable rapid, noninvasive sampling of the intracellular metabolome (for alternative approaches, see Lange et al, 2001; Noh et al, 2007; Villas-Boas and Bruheim, 2007). When spread diffusively so that they cover only a small fraction of the filter surface area, E. coli will double several times on the filter at a growth rate similar to that in liquid media (Yuan et al, 2006). When the agarose-media mixture contained 10 mM ammonium, E. coli grew to an optical density at 650 nm (OD650)∼1.0 with a doubling time of 58 min (Figure 2A, gray triangles).
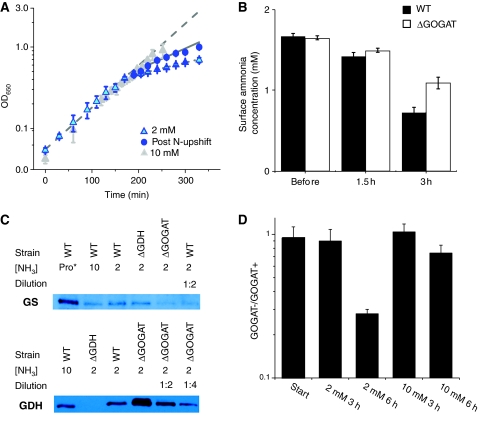
Figure 2.
Filter cultures grown with 2 mM ammonia become nitrogen-limited. (A) Growth of wild-type E. coli with 10 mM ammonium (gray triangles) or 2 mM ammonium (blue triangles) as the nitrogen source. The cells grown in 2 mM ammonium become nitrogen-limited, as indicated by reduced growth rate, after ∼3 h (OD∼0.4). Nitrogen upshift at this point (to 10 mM ammonium plates) partially restores growth rate (blue circles). Error bars show ±s.d. of _N_=3 independent experiments. Lines show fits to exponential growth functions. (B) The ammonia concentration at the surface of the plate becomes depleted coincident with slowing of cellular growth. This depletion is stronger in wild-type cells (black bars) than ΔGOGAT ones (white bars). Error bars show ±s.d. of _N_=3 independent experiments. (C) Western blots of GS (upper) and GDH (lower) in wild-type (WT), ΔGDH, and ΔGOGAT E. coli grown with either proline (pro*), 10 mM ammonia (10) or 2 mM ammonia (2) as the sole nitrogen source. Cells were grown in filter culture on plates to the point of nitrogen limitation (3 h) before sample collection. To obtain a rough estimate of fold over- or under-expression of enzymes in the ΔGOGAT strain, samples were diluted 1:2 or 1:4 in lysis buffer before electrophoresis, as indicated. (D) Competition of ΔlacZ (but otherwise wild type) E. coli with ΔGOGAT E. coli on plates with 2 or 10 mM ammonia as the sole nitrogen source. Error bars show ±s.d. of _N_=4 independent experiments.
To induce nitrogen limitation of filter-grown cultures, we reduced the initial ammonium concentration to 2 mM. The culture grew at the unlimited rate to an OD650∼0.4 (which occurred ∼3 h after transfer of the cells to filter culture), at which time the surface ammonium concentration at the agarose-filter interface became measurably depleted (Figure 2B). As the underlying agarose provides a reservoir of ammonium, growth of the culture did not stop, but continued at a reduced rate for ∼1.5 additional doublings (doubling time ∼220 min) (Figure 2A, cyan triangles). Consistent with the culture being nitrogen-limited, the GS protein content was elevated (Figure 2C). The growth rate was partially restored (doubling time ∼117 min) by transferring the ammonium-limited filter culture to plates with 10 mM ammonium (Figure 2A, blue dots). Thus, although some cell-to-cell variability to ammonia availability likely was present (Atkinson et al, 2002), overall the culture became effectively nitrogen-limited.
Growth in 2 mM initial ammonium concentration was assessed also for the ΔGDH and ΔGOGAT strains. Both grew comparably to wild type (Supplementary Figure 2). Given the higher affinity of the GS/GOGAT pathway than of GDH for ammonia, the lack of a growth defect for the ΔGOGAT strain was surprising, but could be rationalized if growth was limited by ammonium diffusion from the agarose to the filter, rather than by cellular ammonium assimilation. We hypothesized that co-culture should accordingly reveal a growth defect, as the ΔGOGAT strain should be inferior in competing for the limiting ammonium that diffuses to the _E. coli_-filter interface. Indeed, ΔGOGAT was out-competed by the wild-type strain in nitrogen-limited co-cultures (Figure 2D). Consistent with the ΔGOGAT strain being inferior at scavenging ammonium from the plate surface, the surface ammonium concentration at the onset of nitrogen limitation (3 h) was higher for the ΔGOGAT than for the wild-type strain (Figure 2C).
Metabolome changes induced by ammonium upshift center around glutamine and α-ketoglutarate
Having established a technique for ammonium limitation of filter cultures, we measured the dynamic metabolic responses triggered by ammonium upshift (N-upshift). Cells grown on 2 mM ammonium plates to the point of nitrogen limitation (3 h; OD650∼0.4) were transferred to plates with 10 mM ammonium. At various time points thereafter, filters were quenched with cold organic solvent, extracted, and the resulting extracts analyzed by a set of LC-MS/MS methods, which together enable quantitation of ∼250 metabolites (Lu et al, 2008).
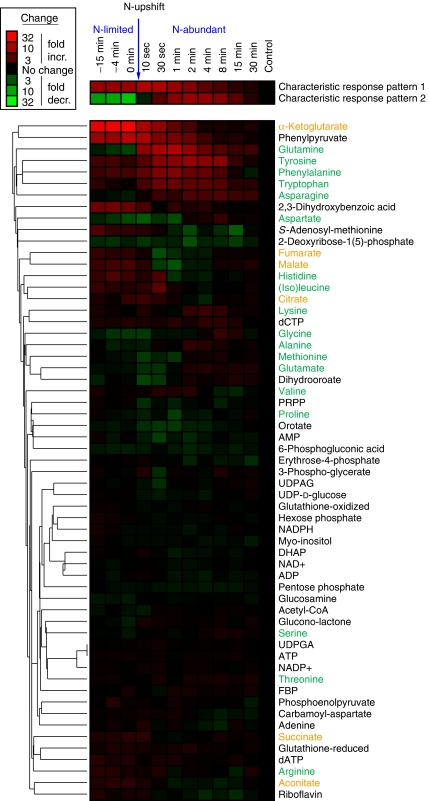
Quantitative data throughout the entire time course were obtained for 59 metabolites (the others were below the limit of quantitation or too unstable to yield reliable information). Dynamics of these 59 compounds (normalized to levels of the corresponding metabolite in exponentially growing E. coli with sufficient ammonium) are shown in Figure 3 in clustered heat-map format. To identify in an unbiased manner the predominant patterns in this data, we used singular value decomposition (SVD) (Alter et al, 2000). The first two characteristic metabolite response patterns obtained by SVD are shown above the heat map. The first characteristic pattern accounts for a remarkable 63% of the overall information in the data, and the second 23%; thus, together, these two patterns account for the majority (86%) of the overall information (Supplementary Figure 3). The compound most strongly contributing to the first characteristic pattern is α-ketoglutarate (Supplementary Table 2), which accumulates dramatically during ammonium limitation and decreases monotonically to a normal level after ammonium upshift. The compound most strongly driving the second characteristic pattern is glutamine, which is depleted during ammonium limitation and transiently overshoots its normal level on nitrogen upshift. Thus, an unbiased data analysis revealed that the compounds whose concentrations are most strongly driving the overall metabolome response are themselves the central players in nitrogen assimilation.
Figure 3.
Metabolome dynamics during ammonia upshift. Columns represent different time points before and after shift from limiting ammonia to 10 mM ammonia. The upshift was applied at _t_=0 min as indicated by the arrow. Fold-change is relative to exponentially growing cells. The data represent the average of _N_=4 independent experiments. The two most characteristic metabolite response patterns, as identified by singular value decomposition (SVD), are shown above the metabolite heat map. Names of amino acids are in green and those of TCA cycle intermediates are in orange.
Other strongly impacted compounds included ones closely tied to α-ketoglutarate and glutamine: other TCA compounds (Figure 3, orange labels) and amino acids (Figure 3, green labels). During ammonium upshift, cofactors such as ATP and NADPH, although reactants in nitrogen assimilation, did not change markedly; neither did the adenylylate energy charge or central carbon compounds outside of the TCA cycle (large dark region at the bottom of Figure 3; Supplementary Figure 4). Thus, short-term concentration responses to nitrogen availability occur in a localized region of metabolism closely related to the limiting nutrient, with homeostasis maintained throughout much of core metabolism.
Qualitative assessment of central nitrogen assimilation
Motivated by the observed metabolome changes revolving around central nitrogen assimilation, we focused on quantitatively modeling central nitrogen assimilation reactions. These efforts were facilitated by having experimentally determined that the key cofactors (ATP, NADPH) remained at homeostasis, thereby enabling approximation of their concentrations as constants.
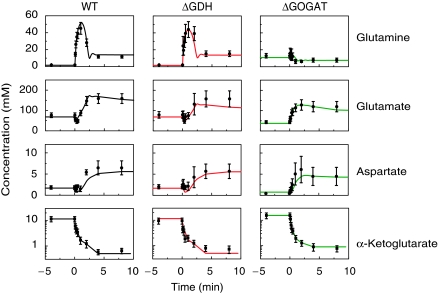
Dynamics of core nitrogen assimilation compounds during nitrogen limitation and subsequent upshift (Figure 4, left column) were qualitatively consistent with expectations based on prior literature: for example, the transient overshoot in glutamine on nitrogen upshift can be explained by high activity of GS in nitrogen-limited cells and subsequent GS inactivation by adenylylation when glutamine accumulates after upshift (Schutt and Holzer, 1972; Kustu et al, 1984). Similarly, the transient dip in glutamate on upshift can be explained by rapid glutamate consumption by GS when ammonium availability increases, followed by increased glutamate production by GOGAT when glutamine accumulates (Miller and Stadtman, 1972; Schutt and Holzer, 1972).
Figure 4.
Measured and modeled dynamics of central nitrogen assimilation intermediates. Symbols represent experimental measurements (_N_=4 for independent experiments for wild type and _N_=2 for ΔGDH and ΔGOGAT). Error bars represent ±s.e., including propagation of error from prior absolute quantitation measurements (see Materials and methods). Lines represent the output of the dynamic model for GLN, GLU, and ASP. For α-ketoglutarate (which was an input to the model), the lines are a multistep linear fit to the data.
A challenge in quantitatively modeling these data is that glutamate can be formed by either the GDH or GS/GOGAT pathways. To eliminate this complexity, we conducted the same ammonium perturbation in ΔGDH and ΔGOGAT E. coli. Consistent with the expectation that the GS/GOGAT pathway dominates under nitrogen limitation, metabolite patterns in the ΔGDH cells were indistinguishable from wild type (Figure 4, middle column), whereas those in ΔGOGAT cells were completely different (Figure 4, right column). Most strikingly, in the ΔGOGAT cells glutamine behavior was opposite to that in wild type. The observed glutamine pattern in the ΔGOGAT cells (elevation during nitrogen limitation and decrease on N-upshift) can be qualitatively rationalized based on lack of glutamate (which falls substantially during nitrogen limitation of ΔGOGAT cells) slowing cellular growth, hence reducing glutamine consumption, and thereby leading to glutamine accumulation.
Another important consideration in quantitatively modeling these data is whether ammonium enters the cell through unfacilitated membrane diffusion or facilitated diffusion through the ammonium channel AmtB. To address this, experiments were conducted in ΔAmtB cells. Consistent with a previous report by Atkinson et al (2002), the growth (data not shown) and metabolome phenotypes of ΔAmtB cells (Supplementary Figure 5) were indistinguishable from wild type under our experimental conditions. As our experiments were conducted at pH∼7, the lack of a phenotype of ΔAmtB is consistent with AmtB being an ammonium cation channel (Fong et al, 2007; Javelle et al, 2008), which enhances cellular access to ammonium primarily at acidic pH, when ammonia (which can pass through the cell membrane by diffusion) is strongly depleted by protonation.
Initial quantitative modeling of central nitrogen assimilation
To translate this qualitative understanding to quantitative equations, we wrote a differential-equation model of the network, including the cascade of covalent modification reactions that controls GS activity through (de)adenylylation (Figure 5). The model treats the extracellular concentration of ammonium and intracellular concentrations of the central carbon metabolites α-ketoglutarate and oxaloacetate as inputs to a nitrogen assimilation module, which converts these metabolites into glutamate, glutamine, and aspartate. The concentrations of glutamine and glutamate control growth rate, which in turn determines the consumption of amino acids to form biomass. The effluxes of amino acids to biomass are a known function of growth rate, dictated by the average composition of E. coli (Reitzer, 2003). These effluxes involve glutamine, glutamate, and aspartate contributing to synthesis of other metabolites (e.g., through transamination), as well as being directly assimilated into protein.
Figure 5.
Elements of the nitrogen assimilation model. The ODE model describes the processes by which ammonia is taken up (N-assimilation) and nitrogen is distributed to biosynthetic pathways (N-utilization). The concentration of species within the dashed boxes (GLU, GLN, ASP, intracellular NH3, and unmodified GS and PII) are calculated as variables by the model, whereas those outside the boxes (α-KG, OA, and extracellular NH3) must be provided as inputs. N-assimilation consists of five metabolic and regulatory reactions: (1) glutamate dehydrogenase, GDH; (2) glutamine synthetase, GS; (3) glutamate synthase, GOGAT; (4) adenylyltransferase/adenylyl-removing enzyme, AT/AR; (5) uridylyltransferase/uridylyl-removing enzyme, UT/UR. Ammonia enters the cell by passive diffusion across the membrane. N-utilization consists of two different classes of reactions, which consume amino acids: those in which a nitrogen group is transferred to an acceptor molecule, recycling the carbon skeleton (N-donation) and those that consume the entire metabolite (protein and biosynthesis). In the model, the rate of GLN, GLU, and ASP consumption for biosynthesis and protein production was taken to be proportional to growth rate (for details, see Supplementary Table 5).
Implementation of the model required defining a variety of parameters: the rate of transport of ammonia into the cell by diffusion (_k_diff), binding affinities of metabolites for enzyme active sites (_K_m and _K_i), maximum enzyme velocities (_V_max), and parameters relating intracellular metabolite concentrations to cellular growth rate. The general philosophy was to take _K_m and _K_i values directly from biochemical literature when available (see Supplementary Table 3), and to select other parameters to fit the observed experimental data shown in Figure 4 using a global search algorithm (genetic algorithm) (Goldberg, 1989; Feng et al, 2006). Values taken from literature were from experiments at physiological pH and temperature whenever possible; nevertheless, these literature values may not precisely reflect enzyme properties in the cellular milieu. The complete set of parameters used in our model is provided in Supplementary Table 3. All parameters were assumed to be constant between the wild-type, ΔGDH, and ΔGOGAT strains, except for the _V_max values for GS and GDH, which varied as a result of differential enzyme concentrations. The relative _V_max values were assumed to be proportional to the associated enzyme protein levels, which were measured directly by western blots (Figure 2C).
Growth rate was simulated as a function of intracellular metabolite concentrations using a variant of a previously described saturable, Hill-type function (Goyal and Wingreen, 2007):
where τ is the doubling time of the cells (i.e., the inverse of growth rate), τ0 is the doubling time with ample ammonium, [_Q_] and [_E_] are the intracellular glutamine and glutamate concentrations, and _K_Q and _K_E are parameters. This formulation satisfies following plausible constraints: τ is a monotonically decreasing function of the two metabolite pools, τ approaches infinity if either pool approaches zero, and τ becomes asymptotically independent of either metabolite pool ([_Q_], [_K_]) above a saturating pool size (_K_Q, _K_E).
The selection of a Hill coefficient of two was based on Ikeda et al's results for Salmonella (Ikeda et al, 1996) and our observation that this choice of Hill coefficient enables us to fit the experimentally observed growth rates based on the experimentally observed glutamine and glutamate concentrations, both before and after nitrogen upshift in wild-type, ΔGDH, and ΔGOGAT E. coli (τ0, _K_Q, and _K_E equal to 90 min, 1.77 mM, and 49.5 mM respectively). The relatively large value of τ0 reflects the fact that the cells, after having experienced nitrogen limitation, have a reduced maximum growth rate (perhaps because of a lower ribosome density; such delayed recovery of growth rate on nitrogen upshift was also observed by Blauwkamp and Ninfa (2002) and has been reported for Salmonella (Kjeldgaard et al, 1958; Bremer and Dennis, 1975)). The value of _K_Q (1.77 mM) was within five-fold of that found by Ikeda et al in Salmonella (0.4 mM) (Ikeda et al, 1996).
Efforts at fitting the dynamic metabolite concentration data using the above framework were, however, initially unsuccessful. In particular, no parameter sets captured the sustained increase in glutamate, after a transient drop, that occurred after ammonium upshift in wild-type and ΔGDH cells (Figure 4).
Competition for the active site of GOGAT controls the glutamate production rate
To assess whether the simulation's failure reflected a logistical difficulty (coding error or inadequacy of the parameter search) or a more fundamental issue, we analyzed glutamate fluxes in ΔGDH cells algebraically, assuming pseudo-steady-state both before and 15 min after the nitrogen upshift. After nitrogen upshift, to enable the ∼2-fold increase in cellular growth rate, glutamate consumption and production must both increase ∼2-fold. The only route of glutamate production in ΔGDH cells is the GOGAT reaction, which is strongly forward driven (ΔG≪0, based on the literature reported ΔG0 and observed metabolite concentrations; Supplementary Table 4A); thus the reverse flux is negligible. Accordingly, the forward rate of this reaction must increase ∼2-fold after nitrogen upshift. Qualitatively, the large increase in cellular glutamine concentration after nitrogen upshift seems adequate to drive this flux increase. Quantitatively, however, the state-steady analysis revealed a problem: even during nitrogen limitation, the cellular glutamine concentration (1.9 mM) is six-fold above the _K_m of GOGAT for glutamine (0.3 mM) (Miller and Stadtman, 1972). Thus, GOGAT is saturated both before and after nitrogen upshift, and the flux through GOGAT should therefore hardly increase even if cellular glutamine concentration increases greatly. Compounding this problem, other metabolite concentration changes that occur after nitrogen upshift (falling α-ketoglutarate and rising glutamate) tend to favor lower, not higher, GOGAT flux.
To quantify the extent of GOGAT saturation by glutamine, we evaluated the elasticity coefficient. By definition, the elasticity coefficient of enzyme flux (V) with respect to changes in substrate or product concentrations (C) is given by ∂ln_V_/∂ln_C_. The elasticity coefficient can be decomposed into a mass action term (which captures the general propensity of substrate addition to favor product formation, and is positive for substrate and negative for product) and a kinetic term (which in our case captures the propensity of both substrate and product to cause enzyme active-site saturation, and is negative for both substrate and product) (Hofmeyr, 1995). As shown in Supplementary Table 4C, for glutamine and GOGAT, the elasticity coefficient was only 0.37 during nitrogen limitation, and yet lower after upshift. These low elasticity coefficients reflected a substantial negative kinetic term indicative of active-site saturation by glutamine, and therefore relative insensitivity of GOGAT flux to glutamine concentration.
Modeling the increase in GOGAT flux after N-upshift required recognition of an important role played by active-site competition. Although product inhibition had been included in the initial failed modeling attempt, competitive inhibition by aspartate, a feature identified indirectly in a classic biochemical study of GOGAT, had not (Miller and Stadtman, 1972). The average occupancy of a particular species X j in a binding site that can accommodate N possible species is
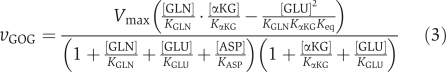
If X j is a substrate, the forward reaction rate is proportional to p j. Thus, the occupancy of GOGAT by glutamine must increase by two-fold for flux to increase two-fold. We find that an adjustment in the Michaelis constant of glutamine (from 0.3 to 0.8 mM) enables this occupancy change while using best estimates of all other metabolite-binding affinities from the literature (Miller and Stadtman, 1972) (Supplementary Tables 3 and 4). The resulting expression for the GOGAT flux is (with the ratio of NADP+ to NADPH, which did not change measurably in response to nitrogen upshift, incorporated into _K_eq)
In equation (3), the numerator reflects the propensity for the reaction to proceed (in the forward minus the backward direction) given no limitation on enzyme active-site availability; the first bracketed term in the denominator reflects competition for the enzyme's glutamine binding site; and the second term reflects competition for the α-ketoglutarate one. Repeating the evaluation of the elasticity coefficient, we find that inclusion of aspartate competition (along with modification of the Michaelis constant of glutamine) roughly triples the elasticity coefficient of GOGAT flux with respect to glutamine concentration (Supplementary Table 4C). In addition, aspartate competition renders the elasticity coefficient of GOGAT flux with respect to glutamate concentration less negative; that is, it mitigates the impact of product inhibition. Both of these changes in elasticity coefficients favor the requisite increase in GOGAT flux after ammonia upshift.
Using the above expression for GOGAT, and similar expressions for all other metabolic reactions (Supplementary Table 5), parameters were identified that resulted in a good fit of the simulation to the experimental data from both wild-type and mutant E. coli (Figure 4).
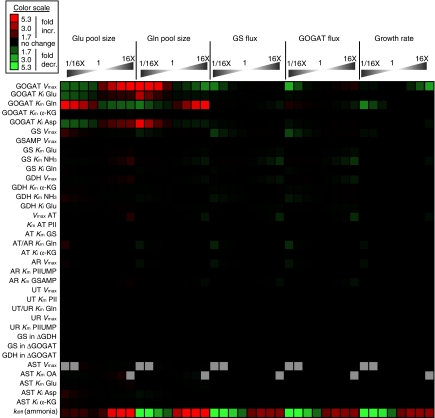
To assess the sensitivity of the model results to the precise values of the parameters, in addition to computing response coefficients (Supplementary Table 7), we ran simulations with each parameter varied by fixed ratios from its optimal value. In the nitrogen-limited regime, the calculated growth rate was insensitive to changes in nearly every enzymatic parameter in the model, with two informative exceptions: growth was impaired by increases to the _V_max of GOGAT, and also by decreases to the Michaelis constant of GOGAT for glutamine (Figure 6). In each case, the glutamine pool is depleted by the increased activity of GOGAT. These predictions highlight the physiological importance of regulation of glutamate production by competition for the GOGAT active site.
Figure 6.
Fluxes and pool sizes under nitrogen limitation were generally robust to changes in model parameters. Simulations were performed in which a single model parameter was multiplied by a factor of (from left to right) 1/16, 1/8, 1/4, 1/2, 2, 4, 8, or 16, and the nitrogen-limited steady-state pool sizes of GLU and GLN, the fluxes in the GS/GOGAT cycle, and the growth rate were calculated. Values shown are ratios between the perturbed simulation and the original. Gray boxes indicate simulations, which did not reach steady state. GOGAT, glutamate synthase; GS, glutamine synthetase; GDH, glutamate dehydrogenase; AT, adenyltransferase; AR, adenyl-removing enzyme; UT, uridyltransferase; UR, uridyl-removing enzyme; AST, aspartate aminotransferase; _k_diff, the diffusion constant for ammonia across the cell membrane. When referring to a _K_m or _K_i value, the substrate is listed after _K_m/i, for example, the _K_m of GOGAT for glutamine is listed GOGAT _K_m Gln.
Control of net aspartate production
To investigate whether active-site competition might be more broadly implicated in regulation of cellular metabolism, we examined also aspartate production from glutamate and oxaloacetate, catalyzed by aspartate aminotransferase (AST). As oxaloacetate was not directly measured because of its low cellular concentration and instability, its concentration was estimated by assuming equilibrium between oxaloacetate and malate (Voet and Voet, 2004).
To feed protein and nucleic acid synthesis, net aspartate production must increase ∼2-fold after nitrogen upshift given the observed two-fold increase in growth rate. This flux increase is not caused simply by increased substrate concentration, as glutamate rises only modestly and oxaloacetate presumably falls with nitrogen upshift. Instead, control of the reverse reaction rate by active-site competition is the most important means of controlling net flux. Before and after nitrogen upshift, AST's two substrate binding sites (one for the four carbon compounds oxaloacetate and aspartate, and the other for the five carbon compounds glutamate and α-ketoglutarate) are both saturated, largely with aspartate and glutamate (Powell and Morrison, 1978; Deu et al, 2002). When these amino acids occupy both sites (the most common situation), no reaction occurs. Thus, the forward and reverse reaction rates are each small compared with the maximum turnover rate of AST. The forward reaction that produces aspartate proceeds only when oxaloacetate is bound and the reverse reaction only when α-ketoglutarate is bound. Of the four reactants and products, the one changing most in concentration with nitrogen upshift is α-ketoglutarate, which decreases ∼20-fold, thereby decreasing by ∼20-fold the rate of the reverse reaction. The net reaction thereby shifts from being weakly forward driven (close to equilibrium) before nitrogen upshift, to largely unidirectional (Supplementary Table 6). This turning off of the reverse reaction results in the requisite increase in net aspartate production on nitrogen upshift (Figure 4).
Evaluating the AST reaction in terms of elasticity coefficients, during nitrogen limitation, the elasticity coefficients of AST flux with respect to substrates and products were high (strongly positive for both substrates and strongly negative for both products; Supplementary Table 6C). These large coefficients, dominated by the mass action terms, reflect the system being near equilibrium, in which a modest change in substrate or product concentrations results in a marked change in net flux. Given the large elasticity coefficients for both substrates and products, the overall flux change was dominated by the metabolite that varied most in concentration after ammonia upshift, α-ketoglutarate.
Predicting the response to other perturbations
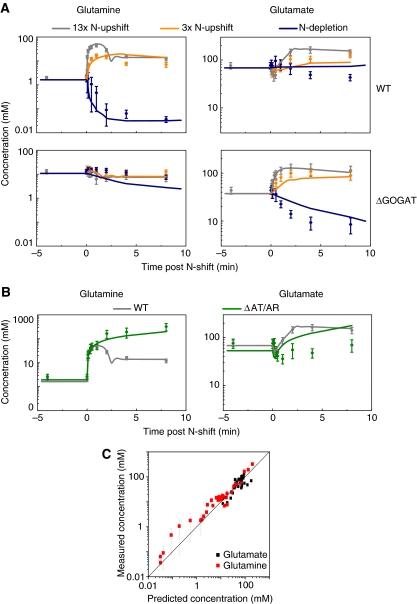
To assess the predictive power of the model, all parameter values were fixed based on the data shown in Figure 4, and subsequently the metabolite profiles for three additional perturbations were simulated and compared with experimental results (Figure 7). The first perturbation involved a nitrogen upshift (the same qualitative perturbation as the training data), but to a lower final ammonium concentration (3-fold upshift instead of 13-fold). The model correctly predicted that, in both wild-type and ΔGOGAT E. coli, the smaller nitrogen upshift (Figure 7A, orange) would result in the same general pattern as the larger upshift (Figure 7A, gray). Moreover, the simulation correctly predicted that the glutamine overshoot and glutamate dip observed in wild type would be reduced in magnitude.
Figure 7.
Comparison of model predictions and experimental observations for (A) additional ammonia perturbations and (B) N-upshift in the ΔAT/AR genetic background; (C) plots the overall extent of agreement. In panels A and B, symbols represent experimental measurements (_N_=4 for independent experiments for wild type and _N_=2 for mutants). Error bars represent ±s.e., including propagation of error from prior absolute quantitation measurements (see Materials and methods). Lines represent the model predictions generated independently from the experimental data. (A) The 13-fold ammonia upshift as per Figure 4 (gray), 3-fold upshift (orange), and N-depletion (dark blue) for the metabolite labeled on top and genetic background on the right. (B) The 13-fold ammonia upshift for wild type (gray) and ΔAT/AR (green), for the metabolite labeled on top. (C) Truth plot showing the correlation between predicted and measured concentrations. Data presented in panels A and B (omitting those in gray, which were used in model training) were pooled to assess the overall predictive power of the model. For each data point, the x axis reflects the model prediction and the y axis the measured concentration. The line is a fit to equation _Y_=X, with _R_=0.85.
To determine whether the model could also predict responses to qualitatively distinct perturbations, we examined a downshift from limiting ammonium to ammonium starvation. Nitrogen-limited cells were transferred to ammonium-free agarose and consumption of the trace residual ammonium on the filters measured (Supplementary Figure 6). These measurements were then used to predict cellular metabolite dynamics (Figure 7A, dark blue). For wild-type cells, glutamine was predicted to fall rapidly, whereas in ΔGOGAT cells glutamine was predicted to hardly fall. In contrast, glutamate was predicted to fall rapidly in ΔGOGAT cells, but not in wild-type cells. These major predictions were experimentally verified, although the quantitative match between the simulations and experimental data was imperfect.
To examine whether the model could also be used to predict responses to genetic perturbations, Δ_glnE_ cells, lacking GS adenyltransferase (AT/AR), were subjected to the 13-fold ammonium upshift. This strain lacks the ability to turn off GS by adenylylation, thereby breaking the feedback circuit that normally limits glutamine accumulation. Results are shown separately from the data for the above perturbations, as the glutamine response would otherwise be off-scale (Figure 7B). The model predicted that glutamine would increase dramatically and persistently with nitrogen upshift, to an order-of-magnitude above the highest concentrations seen in wild type, with the plateau level of glutamine after upshift controlled by direct product inhibition of the GS active site. In contrast, the model predicted that glutamate would closely mimic wild type, showing a transient dip. This prediction for glutamate contrasted with expectations based on prior literature, which suggested that glutamate should be substantially depleted (Kustu et al, 1984). Nevertheless, the model's predictions for both glutamine and glutamate were experimentally verified.
Notably, the ΔAT/AR strain actually grew more slowly after nitrogen upshift than before the onset of N-limitation. Similar impaired growth was reported for ΔAT/AR Salmonella on addition of ammonium to proline-fed cells and was attributed to depletion of the glutamate pool (Kustu et al, 1984). Here, however, we did not observe substantial glutamate depletion (Figure 7B). This discrepancy likely arises from the different N-limiting condition used (proline feeding versus ammonia limitation, which result in the main route to glutamate being different). Instead, the growth defect observed here may be due to excessive glutamine inducing osmotic stress. Consistent with cells' attempting to reduce their internal osmolarity, the ΔAT/AR strain leaked amino acids on nitrogen upshift (Supplementary Figure 7). As the consumption of amino acids by growth was overestimated in the model after N-upshift (because of high glutamine and glutamate concentrations), such unmodeled amino acid leakage likely contributes to the model's ability to predict intracellular amino acid dynamics decently. Such leakage may also contribute to the measured glutamate concentrations being somewhat below those predicted by the simulation.
To assess the predictive power of the model more quantitatively, we plotted model predictions versus experimental results (Figure 7C). To distinguish predictive power from the ability to fit data, all data used for parameter identification were excluded from this analysis. Overall, the ability of the model to predict cellular metabolite concentrations was good, with no evidence of systematic error (95% confidence limits of the slope included 1) and most of the variance explained (_R_=0.85).
Computed phenotypes
Having validated the predictive power of the simulation, we used it to estimate certain intracellular parameters that cannot be readily measured experimentally. One of these is the intracellular ammonium concentration, which the simulation estimates as ∼1 μM in nitrogen-limited wild-type and ΔGDH cells, and ∼30 μM in ΔGOGAT cells. The higher intracellular ammonium concentration in the ΔGOGAT cells accounts for their ability to grow similarly to wild-type cells despite the lower affinity of GDH (_K_m≈1.5 mM) than GS (_K_m≈0.1 mM) for ammonium.
Measuring the dynamic changes in fluxes as a system transitions between steady states remains experimentally infeasible. Results of computational simulation of the central ammonium assimilation fluxes during the transition from limiting to ample ammonium in wild-type E. coli are plotted in Supplementary Figure 8.
Other important parameters that can be readily calculated using the simulation are enzyme flux-control coefficients: the fractional change in pathway flux that is produced by a fractional change in the concentration of the enzyme of interest, that is, (Δ_F_pathway/_F_pathway)/(Δ[_E_j]/[_E_j]) (Fell, 1997). For the nitrogen assimilation pathway (resulting in net ammonium assimilation, glutamate production, and growth), the simulation revealed that, under nitrogen-limiting conditions, the flux-control coefficients of GS, GDH, and GOGAT are negligible, with all flux control resting in diffusion of ammonia into the cell (captured by the parameter _k_diff in the model).
Discussion
E. coli are remarkably efficient at converting nutrient inputs into biomass. This efficiency enables estimation of many of their steady-state metabolic fluxes through linear optimization, an approach called flux balance analysis (FBA) (Edwards et al, 2002). Although FBA is a powerful tool for flux estimation, it does not reveal the chemical events actually controlling metabolite concentrations and fluxes in cells. Here, we attempted to understand quantitatively such regulation, focusing on short-term responses to changes in nitrogen availability.
To this end, we used LC-MS/MS to measure the dynamics of the E. coli metabolome when ammonium availability increases. Large concentration changes in α-ketoglutarate and glutamine, two of the central players in nitrogen assimilation, occurred rapidly (∼10-fold changes in the first minute after upshift). Unbiased statistical identification of the major trends in the metabolome revealed two predominant characteristic response patterns, one mirroring α-ketoglutarate and one mirroring glutamine. Thus, at least among measured metabolites, those most strongly signaling nitrogen availability were species directly involved in nitrogen assimilation. This observation also conforms well to the known regulation by glutamine and α-ketoglutarate of the protein sensors of nitrogen status (e.g., PII) (Ikeda et al, 1996). The α-ketoglutarate response is also interesting in light of evidence that α-ketoglutarate is the principal signal of nitrogen status in cyanobacteria (Forchhammer, 2004).
A substantial fraction of the metabolome (∼27/59) showed a muted version of the α-ketoglutarate and/or glutamine response patterns, with citric acid cycle compounds generally mirroring α-ketoglutarate and amino acids generally mirroring glutamine. Another large set of metabolites hardly changed in concentration (∼30/59). Thus, despite the dramatic alterations to overall cell physiology, which lead to a two-fold increase in growth rate after ammonium upshift, much of the metabolome was effectively insulated from these changes. The homeostatic compounds included ATP and NADPH, both substrates consumed in nitrogen assimilation. Presumably, increased consumption of these compounds on ammonium upshift was offset by increased production.
The observation that co-factors involved in nitrogen assimilation remained homeostatic enabled simulation of the core reactions of nitrogen assimilation as a discrete module, with the measured cellular concentrations of the relevant carbon skeletons (e.g., α-ketoglutarate) considered as inputs to the model. The model included regulation of GS activity by a cascade of covalent modification reactions. In addition to GS modification, the model captured ammonia diffusion into the cell, its assimilation into glutamine, glutamate, and aspartate, and consumption of these species to drive biomass production. Growth rate was simulated as a function of the intracellular concentrations of glutamine and glutamate. The model thereby related environmental conditions to the growth rate through intracellular metabolite levels. To our knowledge, it is the first quantitative model to simulate the chemical steps by which environmental nutrient availability controls, through intracellular metabolite levels, cellular growth.
To the extent possible, the biochemical parameters of the model were taken from literature data. Those parameters not available in the literature (or for which biochemical estimates are generally not indicative of cellular conditions, e.g., _V_max), were determined by using a genetic algorithm to search for parameter values that resulted in the model matching the experimental results. One important conclusion of this integrated experimental-modeling effort was that, for the most part, known regulation of nitrogen assimilation is correct: it was sufficient to result in good agreement between the model and the experimental data.
Perhaps most informative, however, was the exception to this rule: all of the simulations failed to reproduce experimental results unless competition for enzyme active sites was explicitly included. Although theoretical analyses of metabolic regulation have discussed the potential importance of such competition (Fell, 1997), concrete examples of its significance have been lacking. Here we provide two examples. One involves competition of aspartate, glutamate, and glutamine for the active site of GOGAT. Given that neither aspartate nor glutamate is a potent GOGAT inhibitor biochemically (Miller and Stadtman, 1972), such competition was previously overlooked; however, given the high cellular concentrations of these amino acids, this ‘weak' inhibition is nevertheless physiologically critical. Interestingly, aspartate, which is not directly involved in the GOGAT reaction, is a more important active-site inhibitor than glutamate, the enyzme's product. A second example involves control of net aspartate production (by AST), which increases on nitrogen upshift. This net flux increase is achieved not by accelerating the forward reaction but by shutting off the reverse one. The decrease in the reverse flux is accomplished by glutamate out-competing α-ketoglutarate for the enzyme active site.
Our observation that active-site competition has an important function in controlling nitrogen assimilation fluxes matches nicely with the recent finding that most enzymes are substrate saturated in E. coli (Bennett et al, 2009). Both of these findings point to the intracellular milieu being crowded not only with macromolecules (Vazquez et al, 2008), but also with small molecule metabolites, which greatly outnumber macromolecules on a molar basis. Flux control through active-site competition is especially valuable in vivo given such an intracellular environment, because it provides regulation even in the regime in which substrate concentration greatly exceeds the _K_m of the enzyme.
A distinguishing feature of flux control by active-site competition, relative to allostery or protein covalent modification, is that maximum enzyme activity can always be achieved if the substrate pool gets high enough. Thus, while flux control by active-site competition is efficient (not requiring expression of specific regulatory proteins or subunits), it is insufficient when reaching maximum enzyme activity might actually be dangerous to the cell. For example, it is probably critical for cells to be able to shut down GS activity in some circumstances, to save ATP and to prevent excess glutamine production (e.g., in ammonium shock of carbon-starved cells).
This work builds on a rich history of dynamic modeling of biological systems using ordinary differential equations, including in the area of metabolism (Teusink et al, 2000; Rohwer and Botha, 2001; Chassagnole et al, 2002; Snitkin et al, 2008). A distinguishing feature here is integration of systems level experiments with computational modeling to quantitatively understand a metabolic network involving multiple levels of regulation (i.e., protein covalent modification and enzyme active-site competition). We experimentally validated the model's ability to predict not only trends but time-dependent absolute cellular concentrations (Figure 7).
Beyond clarifying mechanisms of cellular metabolic regulation, integrated experimental-modeling studies have the potential to generate models of intrinsic value. One potential value of these models is providing an easy way to determine properties that cannot be readily experimentally measured. Examples include dynamically changing fluxes (Supplementary Figure 8), flux-control coefficients, and sensitivities of metabolite concentrations to enzyme parameters (Figure 6). These properties, together with the measured ones, will then hopefully reveal regulatory principles. As an example, here we propose two principles relating to control of steady-state metabolite concentrations in cells:
- In nutrient-limited cells, the concentration of the growth-limiting metabolite (e.g., glutamine in wild-type cells, glutamate in ΔGOGAT ones) is determined by the sensitivity of growth rate to the metabolite's concentration (i.e., the _K_m of the growth function). The logic is that production of this metabolite is limited by environmental factors (e.g., the amount of ammonium). To balance production with consumption, growth must slow. The concentration in which this slowing occurs determines the metabolite's steady-state value.
- The concentrations of other metabolites are determined by the strength of the feedback control of their production. The logic is that production capacity of most metabolites exceeds demand and is brought into balance by feedback. The concentration in which balance is achieved is controlled by the _K_i (or analogous parameter) of the feedback, as well as the extent of excess production capacity. This concept has been explored in depth mathematically (Hofmeyr and Cornish-Bowden, 2000).
In addition to providing insight into regulation of nitrogen assimilation in particular, and metabolite concentrations and fluxes in general, this work exemplifies the potential for using metabolomic data to drive the development of predictive metabolic models. As parameter identification for large nonlinear dynamic models is unreliable, we believe the most promising route to genome-scale dynamic simulations is through careful development of modular models such as the one presented here. If designed appropriately, these models can then be integrated to yield larger ones. For example, the nitrogen assimilation model, which takes α-ketoglutarate and oxaloacetate concentration as inputs, is well prepared for integration with a modular model of the TCA cycle, which predicts these concentrations, among others. The points in which these modular pathways meet, for example, α-ketoglutarate between the TCA cycle and nitrogen assimilation, should be particularly informative regarding metabolic integration: how information about the availability of one nutrient is communicated to pathways metabolizing others.
Materials and methods
Strains and media
All strains were isogenic of prototrophic E. coli NCM 3722 (Soupene et al, 2003). Strain FG 1088 (Δ_gdhA_∷Kan), FG 1079 (gltD_∷Kan) (Yan, 2007), FG 1114 (Δ_glnE), and NCM 4310 (_amtB_∷Kan) were a generous gift of Dr Dalai Yan, and strain HG 0710 (Δ_lacZ_∷Kan) of Dr Hani Girgis. All strains were cultured in minimal salts media (Gutnick et al, 1969) with 10 mM NH4Cl and 0.4% glucose at 37°C, unless otherwise noted. Plates contained, in addition to the appropriate media, 1.5% ultrapure agarose (triply washed with cartridge-filtered water to remove trace organic contaminants).
Culture conditions and metabolite extraction
Detailed protocols for preparing filter cultures and extracting metabolites have been published (Bennett et al, 2008; Yuan et al, 2008). In brief, filter cultures were prepared by passing 5 ml of exponentially growing liquid batch culture through membrane filters. The filters were then placed on top of agarose loaded with the appropriate minimal media. To measure growth, filters were washed thoroughly with 5 ml of water and absorbance at 650 nm measured. To quench metabolism and initiate extraction, the filters were submerged directly into −75°C methanol or −20°C 40:40:20 acetonitrile:methanol:water with 0.1% formic acid. In both cases, serial extraction (two additional rounds) was used to maximize metabolite yields. All extracts of a given sample were then pooled and mixed with isotope-labeled internal standard compounds (the 10 listed in Bajad et al (2006), as well as isotope-labeled glutamine) and stored at 4°C until analysis. The methanolic extracts were used to generate data on central carbon metabolites and amino acids; the acetonitrile-containing ones for nucleotides and their derivatives (Rabinowitz and Kimball, 2007).
Metabolite measurement
Cell extracts were analyzed by LC-electrospray ionization (ESI)-MS/MS on a Thermo Quantum triple quadrupole mass spectrometer operating in selected reaction monitoring (SRM) mode. All samples were analyzed within 24 h of their preparation. Separate LC runs were conducted for positive and negative ionization modes, using hydrophilic interaction chromatography and ion-pairing reversed phase chromatography, respectively (Lu et al, 2008).
Amino acids were derivatized before their quantitation by LC-ESI-MS/MS. Cell extract (200 μl) was mixed with triethylamine (5 μl) and benzyl chloral formate (1 μl) to convert amines to _N_-benzylcarbamate (Cbz) derivatives (Kraml et al, 2005).
As internal standard signals did not vary substantially or systematically between samples within a given time course, the reported metabolite concentration changes are between-sample ratios of the peak heights of the SRM chromatograms without correction for the internal standard response, except for glutamate and glutamine, in which correction for internal standards was performed to maximize quantitative accuracy.
Flux measurement
Fluxes were measured by kinetic flux profiling (Yuan et al, 2006). Filter cultures were grown on minimal media plates to mid log-phase with 10 mM 14NH4Cl as the sole nitrogen source, and then transferred to minimal media plates containing 10 mM 15NH4Cl as the sole nitrogen source. At various time points (e.g., 10, 30, 60, 120 s) after the transfer, cell extracts were prepared and analyzed by LC-MS/MS. For each metabolite of interest, the multiple isotopomers caused by the 15N-labeling were monitored simultaneously using LC-MS/MS, and fluxes were then calculated based on the kinetics of the replacement of the unlabeled species by the labeled ones. A step-by-step protocol has been published (Yuan et al, 2008).
Ammonium assay
To estimate the ammonium concentration on the surface of a plate, the filter culture was removed and immediately replaced with a piece of fresh preweighed membrane filter. The filter was allowed to absorb media from the surface of the plate for ∼30 s, and then weighed and washed with 1 ml water. The wash was then assayed for ammonium using the indophenol blue method (Aminot et al, 1997), and the concentration of ammonium in the surface of the plate was then calculated. This measurement method captures total NH3+NH4+ content irrespective of protonation state.
Competition assay
Competitions were carried out between the ΔGOGAT strain and a ΔLacZ strain that is otherwise wild type. Each strain was grown separately in liquid batch culture to OD650∼0.1. The strains were then mixed 1:1, loaded onto membrane filters, and allowed to grow on agarose plates with 2 or 10 mM ammonium media. At the indicated time points, cells were washed off filter with minimal media lacking ammonium and analyzed using the Macconkey assay.
Western analysis
Filter-grown cells were washed into water, and the presence of GS and GDH in the resulting cell suspension determined by western. To obtain a rough estimate of fold over- or under-expression of enzymes in the ΔGOGAT strain relative to wild type, samples were diluted 1:2 or 1:4 in lysis buffer before electrophoresis. GS antibody was purchased from Abcam (Cambridge, MA). GDH antibody was a gift from D Yan.
Ammonium perturbation time-course experiment
For all ammonium perturbation time courses, filter cultures were grown on minimal media plates with 2 mM NH4Cl as sole nitrogen source for 3 h (the point of initial ammonium limitation), and then transferred to plates with the desired postperturbation ammonium concentration (10 mM NH4Cl for the 13-fold upshift, 2 mM NH4Cl for the 3-fold upshift, and no ammonium for downshift). At various time points preceding (−15, −4, 0 min) and following (5, 10, 20, 30 s, 1, 2, 4, 8, 15, 30 min) the perturbation, metabolism was quenched and metabolites extracted as described above.
All data presented are averages of multiple independent experiments conducted on separate days. On each day, two replicate cultures of exponentially growing cells were analyzed. For each metabolite, the average signal in these samples (_S_exp) was used to determine a conversion factor between measured ion counts in samples taken on that experimental day (S j) and absolute concentration of the metabolite, using the previously determined absolute concentration of the metabolite in exponentially growing cells (_C_exp) (Bennett et al, 2009):
where R _j_=S j/_S_exp and C j is the absolute metabolite concentration in sample j.
Values of C j were then averaged across the multiple independent experiments on different days to obtain C̄ j. Standard error (s.e.) in C̄ j (Δ_C_ j) was determined by standard propagation of error:
where Δ_R_ j is the standard error of the replicate measurements of R j made on different days, R̄ j is the average measured value of R, and Δ_C_exp is the s.e. of C based on prior measurements (Bennett et al, 2009).
Data taken at −15, −8, −4, and 0 min did not differ significantly and were accordingly averaged to estimate the preperturbation steady-state metabolite levels. The primary mass spec data can be downloaded from https://ProteomeCommons.org with following hash code: kiaAkSlMliPpPPX0irpSWGh6ZjUsxxCfn75I8qJrk3e13k5O3FWEHyKt0BbDP+Q7B TVZ4W+8HnASbTe4HOSnfrYMx00AAAAAAAGxVw==
Data analysis
Metabolite-concentration changes (expressed as log2 ratios) were clustered based on Euclidean distance between metabolites using software available through the Princeton Microarray Database (http://puma.princeton.edu). SVD analysis was performed using similarly accessed software.
Model formulation
Differential equations describing the time-dependent concentrations of glutamate, glutamine, aspartate, intracellular ammonium, and the average modification states of GS and PII were formulated assuming reversible Michaelis–Menten kinetics (Supplementary Table 5). Equilibrium constants were taken from Bruggeman et al and adjusted to account for the measured (and time invariant) intracellular ratios ([ADP] × [Pi])/[ATP] and [NADP+]/[NADPH] when appropriate. The rate of passive diffusion of ammonia across the membrane was approximated as _k_diff([NH3]ext−[NH3]int). The model requires as inputs the experimentally observed time-dependent intracellular concentrations of α-ketoglutarate and malate, and the measured extracellular ammonium concentration. The model estimates these concentrations between measured time points through linear interpolation. As significant depletion of surface ammonium was not observed over the 30 min period of interest after upshift, the ammonium concentration after upshift was taken as the relevant media concentration (10 or 2 mM).
Parameter identification and simulation
In vitro estimates of _K_m and _K_i values for GS, GDH, GOGAT, and AST were taken from the literature when available (Supplementary Table 3). The concentration of GS and GDH in the ΔGOGAT strain relative to wild type was estimated from the western analysis. All other parameters were estimated by a genetic algorithm (Feng et al, 2006), which searched for parameter sets that minimized the mean square error between simulated and observed metabolite concentrations, with data points weighted according to the inverse of the experimental error (i.e., the smaller the observed error, the greater the cost of the model deviating from the observed experimental mean). Simulated time courses were obtained by numerically integrating the system of differential equations. A representative parameter set was used to generate the simulation results shown in Figures 4, 6, 7, and Supplementary Figure 8. Although some parameter values were not tightly constrained, simulation results were similar for all of the best-scoring parameter sets generated by the genetic algorithm. The dichotomy between diverse parameter values and tight simulation results in metabolic models has been described earlier (Gutenkunst et al, 2007; Piazza et al, 2008). An SBML format of the model is included in the Supplementary information. All simulation and parameter search code was originally written in C++ and is available on request.
Sensitivity analysis
Simulations were conducted in which one parameter was increased or decreased by a power of 2 from its value in the standard set, up to a maximum of 16-fold in either direction. The simulation was then allowed to reach nitrogen-limited steady state (i.e., equivalent to _t_=0 in Figures 3, 4, and 7).
Supplementary Material
Supplemental figures and tables
Supplementary figures S1–8, Supplementary tables S1–7
code for matlab simulation
Code for matlab simulation
Dataset
Master raw data file
Acknowledgments
We thank Sydney Kustu, David Botstein, and Sidhartha Goyal for their intellectual contributions; Xiaobo Ke for his experimental contributions; Dalai Yan for the ΔGDH, ΔGOGAT, ΔAT/AR strains, GDH antibody, and discussions; and Hani Girgis for the ΔlacZ strain. This work was funded by National Science Foundation CAREER Award MCB-0643859 and Beckman Foundation and American Heart Association Awards to JDR and the National Institute of General Medical Sciences Center for Quantitative Biology/National Institutes of Health Grant P50 GM-071508 and USEPA STAR Grant GAD R 832721-010.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alibhai M, Villafranca JJ (1994) Kinetic and mutagenic studies of the role of the active site residues Asp-50 and Glu-327 of Escherichia coli glutamine synthetase. Biochemistry 33: 682–686 [DOI] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D (2000) Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA 97: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminot A, Kirkwood DS, Kerouel R (1997) Determination of ammonia in seawater by the indophenol-blue method: evaluation of the ICES NUTS I/C 5 questionnaire. Marine Chem 56: 59–75 [Google Scholar]
- Atkinson MR, Blauwkamp TA, Bondarenko V, Studitsky V, Ninfa AJ (2002) Activation of the glnA, glnK, and nac promoters as Escherichia coli undergoes the transition from nitrogen excess growth to nitrogen starvation. J Bacteriol 184: 5358–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD (2006) Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A 1125: 76–88 [DOI] [PubMed] [Google Scholar]
- Bancroft S, Rhee SG, Neumann C, Kustu S (1978) Mutations that alter the covalent modification of glutamine synthetase in Salmonella typhimurium. J Bacteriol 134: 1046–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B, Yuan J, Kimball E, Rabinowitz J (2008) Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc 3: 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BD, Kimball E, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature Chem Biol 5: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauwkamp TA, Ninfa AJ (2002) Physiological role of the GlnK signal transduction protein of Escherichia coli: survival of nitrogen starvation. Mol Microbiol 46: 203–214 [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Yuan J, Bennett BD, Lu W, Kimball E, Botstein D, Rabinowitz JD (2006) Conservation of the metabolomic response to starvation across two divergent microbes. Proc Natl Acad Sci USA 103: 19302–19307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H, Dennis PP (1975) Transition period following a nutritional shift-up in the bacterium Escherichia coli B/r: stable RNA and protein synthesis. J Theor Biol 52: 365–382 [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, Boogerd FC, Westerhoff HV (2005) The multifarious short-term regulation of ammonium assimilation of Escherichia coli: dissection using an in silico replica. FEBS J 272: 1965–1985 [DOI] [PubMed] [Google Scholar]
- Chassagnole C, Noisommit-Rizzi N, Schmid JW, Mauch K, Reuss M (2002) Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol Bioeng 79: 53–73 [DOI] [PubMed] [Google Scholar]
- Deu E, Koch KA, Kirsch JF (2002) The role of the conserved Lys68*:Glu265 intersubunit salt bridge in aspartate aminotransferase kinetics: multiple forced covariant amino acid substitutions in natural variants. Protein Sci 11: 1062–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Covert M, Palsson B (2002) Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol 4: 133–140 [DOI] [PubMed] [Google Scholar]
- Fell D (1997) Understanding the Control of Metabolism. London: Portland Press [Google Scholar]
- Feng XJ, Rabitz H, Turinici G, Le Bris C (2006) A closed-loop identification protocol for nonlinear dynamical systems. J Phys Chem A 110: 7755–7762 [DOI] [PubMed] [Google Scholar]
- Fong RN, Kim KS, Yoshihara C, Inwood WB, Kustu S (2007) The W148L substitution in the Escherichia coli ammonium channel AmtB increases flux and indicates that the substrate is an ion. Proc Natl Acad Sci USA 104: 18706–18711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K (2004) Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev 28: 319–333 [DOI] [PubMed] [Google Scholar]
- Garcia E, Rhee SG (1983) Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII uridylyltransferase and uridylyl-removing enzyme. J Biol Chem 258: 2246–2253 [PubMed] [Google Scholar]
- Goldberg DE (1989) Genetic Algorithms in Search, Optimization, and Machine Learning. Toronto, ON, Canada: Addison-Wesley Longman Publishing Co, Inc. [Google Scholar]
- Goyal S, Wingreen NS (2007) Growth-induced instability in metabolic networks. Phys Rev Lett 98: 138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP (2007) Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol 3: 1871–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutnick D, Calvo JM, Klopotowski T, Ames BN (1969) Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J Bacteriol 100: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyaneshwar P, Paliy O, McAuliffe J, Popham DL, Jordan MI, Kustu S (2005) Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J Bacteriol 187: 1074–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich R, Schuster S (1996) The Regulation of Cellular Systems. New York: Chapman and Hall [Google Scholar]
- Hofmeyr JH (1995) Metabolic regulation: a control analytic perspective. J Bioenerg Biomembr 27: 479–490 [DOI] [PubMed] [Google Scholar]
- Hofmeyr JS, Cornish-Bowden A (2000) Regulating the cellular economy of supply and demand. FEBS Lett 476: 47–51 [DOI] [PubMed] [Google Scholar]
- Ikeda TP, Shauger AE, Kustu S (1996) Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J Mol Biol 259: 589–607 [DOI] [PubMed] [Google Scholar]
- Javelle A, Lupo D, Ripoche P, Fulford T, Merrick M, Winkler FK (2008) Substrate binding, deprotonation, and, selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB. Proc Natl Acad SciUSA 105: 5040–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Mayo AE, Ninfa AJ (2007) Escherichia coli glutamine synthetase adenylyltransferase (ATase, EC 2.7.7.49): kinetic characterization of regulation by PII, PII-UMP, glutamine, and alpha-ketoglutarate. Biochemistry 46: 4133–4146 [DOI] [PubMed] [Google Scholar]
- Jiang P, Ninfa AJ (2007) Escherichia coli PII signal transduction protein controlling nitrogen assimilation acts as a sensor of adenylate energy charge in vitro. Biochemistry 46: 12979–12996 [DOI] [PubMed] [Google Scholar]
- Jiang P, Peliska JA, Ninfa AJ (1998a) Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37: 12782–12794 [DOI] [PubMed] [Google Scholar]
- Jiang P, Peliska JA, Ninfa AJ (1998b) The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry 37: 12802–12810 [DOI] [PubMed] [Google Scholar]
- Kjeldgaard N, Maaløe O, Schaechter M (1958) The transition between different physiological states during balanced growth of Salmonella typhimurium. J General Microbiol 19: 607. [DOI] [PubMed] [Google Scholar]
- Kraml CM, Zhou D, Byrne N, McConnell O (2005) Enhanced chromatographic resolution of amine enantiomers as carbobenzyloxy derivatives in high-performance liquid chromatography and supercritical fluid chromatography. J Chromatogr A 1100: 108–115 [DOI] [PubMed] [Google Scholar]
- Kustu S, Hirschman J, Burton D, Jelesko J, Meeks JC (1984) Covalent modification of bacterial glutamine synthetase: physiological significance. Mol Gen Genet 197: 309–317 [DOI] [PubMed] [Google Scholar]
- Lange HC, Eman M, van Zuijlen G, Visser D, van Dam JC, Frank J, de Mattos MJ, Heijnen JJ (2001) Improved rapid sampling for in vivo kinetics of intracellular metabolites in Saccharomyces cerevisiae. Biotechnol Bioeng 75: 406–415 [DOI] [PubMed] [Google Scholar]
- Lu W, Bennett BD, Rabinowitz JD (2008) Analytical strategies for LC-MS-based targeted metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci 871: 236–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantsala P, Zalkin H (1976) Properties of apoglutamate synthase and comparison with glutamate dehydrogenase. J Biol Chem 251: 3300–3305 [PubMed] [Google Scholar]
- Miller RE, Stadtman ER (1972) Glutamate synthase from Escherichia coli. An iron-sulfide flavoprotein. J Biol Chem 247: 7407–7419 [PubMed] [Google Scholar]
- Ninfa AJ, Atkinson MR (2000) PII signal transduction proteins. Trends Microbiol 8: 172–179 [DOI] [PubMed] [Google Scholar]
- Ninfa AJ, Jiang P (2005) PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol 8: 168–173 [DOI] [PubMed] [Google Scholar]
- Noh K, Gronke K, Luo B, Takors R, Oldiges M, Wiechert W (2007) Metabolic flux analysis at ultra short time scale: isotopically non-stationary 13C labeling experiments. J Biotechnol 129: 249–267 [DOI] [PubMed] [Google Scholar]
- Piazza M, Feng XJ, Rabinowitz JD, Rabitz H (2008) Diverse metabolic model parameters generate similar methionine cycle dynamics. J Theor Biol 251: 628–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JT, Morrison JF (1978) The purification and properties of the aspartate aminotransferase and aromatic-amino-acid aminotransferase from Escherichia coli. Eur J Biochem 87: 391–400 [DOI] [PubMed] [Google Scholar]
- Rabinowitz JD, Kimball E (2007) Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal Chem 79: 6167–6173 [DOI] [PubMed] [Google Scholar]
- Reitzer L (2003) Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol 57: 155–176 [DOI] [PubMed] [Google Scholar]
- Rohwer JM, Botha FC (2001) Analysis of sucrose accumulation in the sugar cane culm on the basis of in vitro kinetic data. Biochem J 358: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto N, Kotre AM, Savageau MA (1975) Glutamate dehydrogenase from Escherichia coli: purification and properties. J Bacteriol 124: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt H, Holzer H (1972) Biological function of the ammonia-induced inactivation of glutamine synthetase in Escherichia coli. Eur J Biochem 26: 68–72 [DOI] [PubMed] [Google Scholar]
- Senior PJ (1975) Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous-culture technique. J Bacteriol 123: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snitkin ES, Dudley AM, Janse DM, Wong K, Church GM, Segre D (2008) Model-driven analysis of experimentally determined growth phenotypes for 465 yeast gene deletion mutants under 16 different conditions. Genome Biol 9: R140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorribas A, Savageau MA (1989) A comparison of variant theories of intact biochemical systems. II. Flux-oriented and metabolic control theories. Math Biosci 94: 195–238 [DOI] [PubMed] [Google Scholar]
- Soupene E, van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, Lee H, Prasad G, Paliy O, Charernnoppakul P, Kustu S (2003) Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J Bacteriol 185: 5611–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teusink B, Passarge J, Reijenga CA, Esgalhado E, van der Weijden CC, Schepper M, Walsh MC, Bakker BM, van Dam K, Westerhoff HV, Snoep JL (2000) Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267: 5313–5329 [DOI] [PubMed] [Google Scholar]
- Vazquez A, Beg QK, Demenezes MA, Ernst J, Bar-Joseph Z, Barabasi AL, Boros LG, Oltvai ZN (2008) Impact of the solvent capacity constraint on E. coli metabolism. BMC Syst Biol 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villas-Boas SG, Bruheim P (2007) Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal Biochem 370: 87–97 [DOI] [PubMed] [Google Scholar]
- Voet D, Voet JG (2004) Table 21-2. Standard free energy changes and physiological free energy changes of citric acid cycle reactions.In Biochemistry, p 790 New York: J. Wiley & Sons [Google Scholar]
- Yan D (2007) Protection of the glutamate pool concentration in enteric bacteria. Proc Natl Acad Sci USA 104: 9475–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Bennett B, Rabinowitz J (2008) Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nature Protoc 3: 1328–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Fowler WU, Kimball E, Lu W, Rabinowitz JD (2006) Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat Chem Biol 2: 529–530 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures and tables
Supplementary figures S1–8, Supplementary tables S1–7
code for matlab simulation
Code for matlab simulation
Dataset
Master raw data file