Three-dimensional collagen represses cyclin E1 via beta one integrin in invasive breast cancer cells (original) (raw)
. Author manuscript; available in PMC: 2013 Apr 25.
Published in final edited form as: Breast Cancer Res Treat. 2010 Jul 4;127(2):397–406. doi: 10.1007/s10549-010-1013-x
Abstract
The behavior of breast epithelial cells is influenced by their microenvironment, which includes stromal cells and extracellular matrix (ECM). During cancer progression, the tissue microenvironment fails to control proliferation and differentiation, resulting in uncontrolled growth and invasion. Upon invasion, the ECM encountered by breast cancer cells changes from primarily laminin and collagen IV to primarily collagen I. We show here that culturing invasive breast cancer cells in 3-dimensional (3D) collagen I inhibits proliferation through direct regulation of cyclin E1, a G1/S regulator that is overexpressed in breast cancer. When the breast cancer cell line MDA-MB-231 was cultured within 3D collagen I gels, the G1/S transition was inhibited as compared to cells cultured on conventional 2D collagen or plastic dishes. Cells in 3D collagen downregulated cyclin E1 protein and mRNA, with no change in cyclin D1 level. Cyclin D1 was primarily cytoplasmic in 3D cultures and this was accompanied by decreased phosphorylation of Rb, a nuclear target for both cyclin E1- and cyclin D1-associated kinases. Positive regulators of cyclin E1 expression, the transcription factor c-Myc and cold-inducible RNA binding protein (CIRP), were decreased in 3D collagen cultures, while the collagen I receptor β1 integrin was greatly increased. Inhibition of β1 integrin function rescued proliferation and cyclin E1 expression as well as c-Myc expression and Rb phosphorylation, but cyclin D1 remained cytoplasmic. We conclude that cyclin E1 is repressed independent of effects on cyclin D1 in a 3-dimensional collagen environment and dependent on β1 integrin interaction with collagen I, reducing proliferation of invasive breast cancer cells.
Keywords: 3-dimensional collagen, breast cancer, cyclin E1, cell cycle, β1 integrin
Introduction
Breast cancer is the most common cancer in American women and remains second only to lung cancer as the leading cause of cancer-related death among women. The information provided from traditional prognostic factors such as age, lymph node status or tumor grade is often not precise enough to tailor cancer diagnosis and treatment effectively. A better understanding of breast cancer biology and identification of accurate prognostic indicators and predictors of response based on the underlying molecular biology could profoundly decrease metastatic death [1, 2].
One important aspect of cancer biology is uncontrolled cell proliferation. Events affecting checkpoints that govern transit through the first gap phase (G1 phase) of the cell cycle are observed frequently in human cancer. Cells are responsive to extracellular signals at G1 phase to decide if cell division continues. Cyclin E1, as regulator of re-entry into G1 from G0 [3] and the G1 to S phase (DNA synthesis) transition [4], has been identified as a prognostic factor in breast cancer. Cyclin E1 is overexpressed in about 32% of breast tumors [5] where it is strongly predictive of poor patient outcome and has been implicated directly in breast cancer etiology [6-8]. Overall, a more detailed understanding of cyclin E1 function in breast cancer biology is needed in order to evaluate its potential as a therapeutic target, prognostic marker and/or predictive marker.
Mammary epithelial (ME) cells respond to signals from the extracellular matrix (ECM) by proliferating and differentiating into highly organized and polarized glands, but in neoplastic progression, the microenvironment is altered such that controls from the ECM are lost. The ECM primarily controls the expression of D-cyclins in early G1 phase; however, cyclin E1 is also responsive to extracellular signals [9-11]. α2β1 integrin, a receptor for collagen I and laminin, can stimulate expression of cyclin E1 and cdk2 as well as cyclin E1/cdk2 complex formation in ME cells in a collagen I-dependent and cyclin D1-independent manner [12]. Based on this and the fact that breast cancer cells directly encounter collagen I during invasion once the basement membrane is compromised [13, 14], we set out to determine how a 3-dimensional (3D) environment composed of collagen I regulates cyclin E1.
The invasive ME adenocarcinoma cell line MDA-MB-231 was cultured either in conventional 2D monolayer on plastic or collagen I, or within a 3D collagen gel to mimic the environment that invasive breast cancer cells encounter as pericellular proteolysis begins. Our results demonstrate that β1 integrin can inhibit the breast cancer cell cycle in a 3D collagen environment via negative regulation of cyclin E1, a very different outcome as compared to a 2D collagen environment.
Material and methods
Cell Culture
The human ME adenocarcinoma cell line MDA-MB-231 (HTB-26, <passage 20) and nontumorigenic human ME cell line MCF10A (CRL-10317, <passage 10) were obtained from American Type Culture Collection (ATCC, Manassas, VA), as were MCF-7, Hs-578T, and SK-BR3 cells. Collagen I was prepared from rat tails using established methods [15] and assessed for polymerization and cell viability. Cells were cultured in 2D monolayer on plastic dishes (2DP), 2D monolayer on plastic dishes coated with a thin (< 0.5mm) layer of 2.5% collagen I (2DCI), or 3D culture within intact collagen I (3DCI, 0.5cm). 3D collagen cultures were prepared as described with modifications [16]. Collagen gelation was induced by incubating half of the collagen solution at 37°C for 1h. Cells were suspended with the other half of liquid collagen and spread on top of the gelled layer. After incubation at 37°C for 1h, complete medium was added. Medium was changed every 2 days and cells were collected at 70-80% confluency for experiments.
Cell Cycle Analysis
For analysis of asynchronous cells, cells were grown for 48h. Cells from 2D cultures were trypsinized and resuspended in complete media followed by brief centrifugation. 3D collagen cultures were treated with 100U/ml highly purified collagenase (Collagenase VII, Sigma) for 20-30min at 37°C. Cell pellets from the three culture conditions were analyzed by flow cytometry using Becton Dickinson FACScan (San Jose, CA) and CellQuest software. For analysis of synchronized cells, cells grown on 2DP and 3DCI to 50%-60% confluence were synchronized in G1 by serum deprivation for 48h followed by 24h treatment with 5μg/ml aphidicolin. Synchronized cells were stimulated with serum and harvested at the indicated times. The experiments were repeated at least 3 times, unless otherwise noted in the figure legend.
Western Analysis
Whole-cell lysates were resolved by 10% SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked with 5% non-fat milk in TBS-0.05% TWEEN20. Primary antibodies against cyclin E1, cyclin D1, HuR, and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA); CIRP (Proteintech Group, Inc., Chicago, IL); Phospho-Rb(Ser780), total Rb and c-Myc (Cell Signaling Technology, Danvers, MA); and β1-integrin (BD Biosciences, San Jose, CA) were used. Blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL). Images were captured using Kodak Image Station 4000MM (Kodak Molecular Imaging) and intensities were quantified by KODAK Molecular Imaging Software v4.0. All experiments were repeated at least 3 times, unless otherwise noted in the figure legend.
Real-Time PCR
Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA). Reverse transcription was performed using oligo(dT)12-18 or random hexamers (Invitrogen).. cDNAs were amplified by real-time PCR using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). Relative quantification standard real-time PCR was carried out using an AB 7500 Fast Real-Time PCR System with these conditions: 10min at 95°C, 40 cycles of 15sec at 95°C, 1min at 60°C. Primers were: cyclin E1 and GAPDH as previously described [17]; 18s rRNA, forward 5′-CGAACGTCTGCCCTATCAACTT-3′ and reverse 5′-ACCCGTGGTCACCATGGTA-3′. Threshold cycles (Ct) were normalized to GAPDH or 18s rRNA. Data were analyzed by Sequence Detection Software 1.31.22 (Applied Biosystems). For mRNA stability assays, cells were treated with 5μg/ml actinomycin D, total RNA extracted at the indicated time following treatment, and real-time PCR performed to determine mRNA half-life. Experiments were performed 3 times.
Immunofluorescence Analysis
Cells grown on glass coverslips in 24-well plates to 70% confluency [16] were incubated with primary antibodies against cyclin D1, CIRP, HuR, or Ki-67 (BD Biosciences) and Alexa Fluor 488 conjugated secondary antibodies. Coverslips were mounted onto slides with VECTASHIELD containing DAPI (Vector laboratories, Inc, Burlingame, CA). Slides were analyzed using a Zeiss Axiovert 200M or a Zeiss LSM 510 Meta confocal microscope and associated software. Experiments were repeated a minimum of 3 times.
β1 Integrin Function Blocking Assay
MDA-MB-231 cells grown in 3D collagen for 24h were treated with media containing AIIB2, a β1 integrin function-blocking antibody [18], for 2h and then collected for Western or immunofluorescence analysis. AIIB2 (Developmental Studies Hybridoma Bank, Iowa City, IA), or IgG1 as a control were used at 0.24mg/ml as this concentration efficiently blocked β1 integrin function in MDA-MB-231 cells grown in Matrigel [19]. Experiment was performed a minimum of 3 times.
Results
3D collagen culture inhibits cell cycle progression
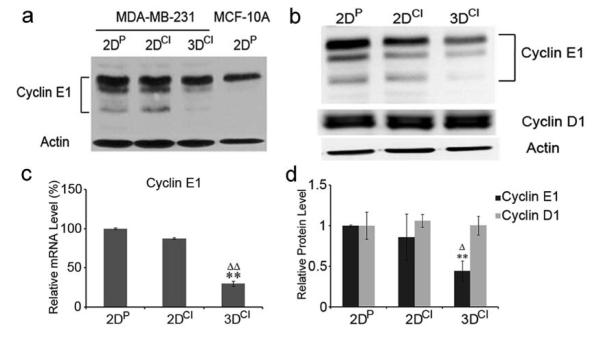
To test if the ECM can control ME cell proliferation through direct regulation of cyclin E1, MDA-MB-231 breast cancer cells were cultured in 2D monolayer on plastic or collagen I, or within 3D collagen gels. We chose MDA-MB-231 cells because cyclin E1 is highly expressed in these estrogen-receptor negative, invasive cancer cells as compared to nontumorigenic ME cells, such as MCF10A [20]. While MCF10A cells express only full-length cyclin E1, MDA-MB-231 cells express both full length and low molecular weight (LMW) isoforms, consistent with their tumorigenicity ([21], Fig. 2a).
Fig. 2.
3D collagen culture downregulates cyclin E1 without changing cyclin D1. a Western blot of cyclin E1 in 2DP, 2DCI and 3DCI MDA-MB-231 cells and 2DP MCF-10A cells. β-actin was used as a loading control. Representative of 2 experiments. b Western blots of cyclin E1 and cyclin D1 in 2DP, 2DCI and 3DCI MDA-MB-231 cells. The relative quantity of cyclin E1 and cyclin D1 was calculated after normalization to actin (d) , expressed as mean ± SD and assessed by Student’s t test. Δp < 0.05 vs. 2DCI , **p < 0.01 vs. 2DP. c Cyclin E1 mRNA level was determined by real-time RT-PCR. Threshold cycles (_C_t values) were normalized to GAPDH or 18s rRNA and plotted as relative mRNA levels. Values were expressed as mean ± SD and assessed by Student’s t test. **p < 0.01 vs. 2DP, ΔΔp < 0.01 vs. 2DCI
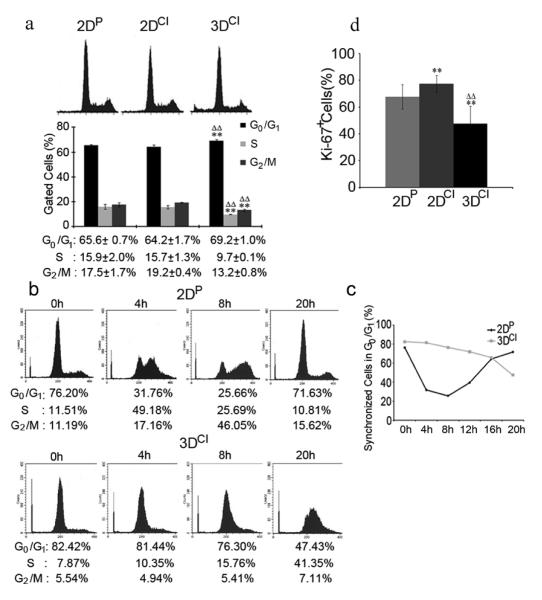
Cell cycle phase distribution was obtained by flow cytometry of asynchronized cells as shown in Fig. 1a. While 2D monolayer cells had a similar cell cycle profile, in 3D collagen G1 phase cells increased by 5% while S and G2/M cells both decreased by 6%. There was also a significant increase in hypodiploid cells in 3D collagen (7.8%) as compared to 2D cultures (1%) indicating increased cell death. This was further confirmed by DAPI staining showing condensed and fragmented nuclei characteristic of apoptosis (data not shown). Cell cycle profile was also analyzed in cells synchronized at the G1/S border. Cells on 2D plastic were compared to cells in 3D collagen since 2D asynchronized cultures showed similar cell cycle profiles. Synchronized cells on 2D plastic completed an entire cell cycle within 20h after serum stimulation (Fig. 1b and c). In 3D collagen, there was a delay entering S phase (Fig. 1b and c). While 49% of cells cultured in monolayer had entered S phase by 4h following serum stimulation, only 10% of cells cultured in 3D had done so. By 20h, 47% of 3D cells were still in G0/G1 while 41% were in S phase. Surprisingly, up to 72 hours after serum stimulation the majority of 3D cells (45%) were still stagnant in G1/S and 14% of cells were hypodiploid (data not shown). These data showed that 3D collagen I significantly delayed the G1/S phase transition in synchronized cells.
Fig. 1.
3D collagen culture inhibits cell cycle progression. a Cell cycle analysis of asynchronous MDA-MB-231 cells in 2DP, 2DCI, or 3DCI. Cell cycle phase distribution was obtained and percentage of gated cells in each phase were expressed as mean ± SD and assessed by Student’s t test; **p < 0.01 vs. 2DP, ΔΔp < 0.01 vs. 2DCI. b Cell cycle analysis of synchronized MDA-MB-231 cells on 2DP or in 3DCI. Data is representative of 2 experiments. c Percentage of synchronized cells in G0/G1 from B was plotted for the indicated time points. d Quantitative analysis of proliferation in asynchronous cells. Percentage of Ki-67+ cells was calculated by counting at least 10 40X fields (at least 300 cells) per condition. **p < 0.01 vs. 2DP, ΔΔp < 0.01 vs. 2DCI
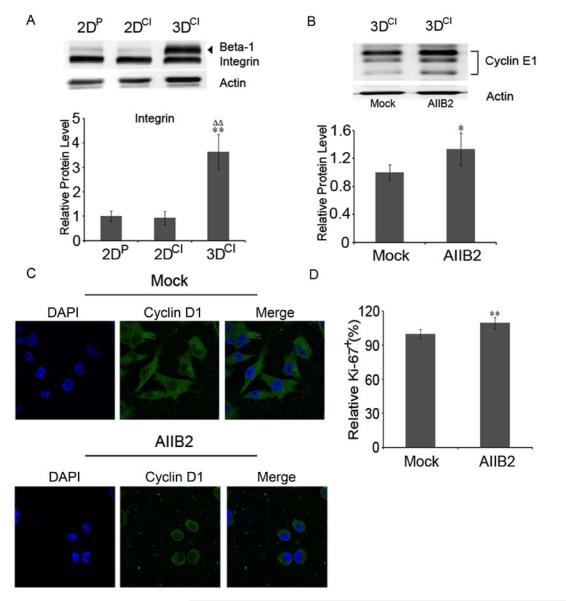
Immunofluorescence analysis of Ki-67, a nuclear protein expressed by cells in all phases of the cell cycle but absent in resting (G0) cells, was used to assess proliferation index in the 3 culture conditions. Fig. 1d shows that 68% of cells on 2D plastic, 77% of cells on 2D collagen and only 48% of cells in 3D were Ki-67+. These data suggest that a 3D environment composed of collagen I suppressed cell cycle progression by delaying entry into S phase and increasing the G0 population.
3D collagen downregulates cyclin E1 without changing cyclin D1
Since a significant G1/S delay was observed in cells cultured in 3D collagen and cyclin E1 functions in the G1/S transition, we first examined potential effects on cyclin E1. The relative level of cyclin E1 (full length and LMW isoforms) was determined by western blotting and quantitated after normalizing to β-actin (Fig. 2b). Cyclin E1 was decreased by 56% in 3D collagen compared to 2D plastic (p<0.01) and 42% compared to 2D collagen (p<0.05), with no significant difference between 2D cultures. Consistent with these results, real time RT-PCR showed that there was no significant difference in cyclin E1 mRNA level between 2D cultures, while cyclin E1 mRNA was decreased by 71% in 3D compared to 2D plastic (p<0.01) and 59% compared to 2D collagen (p<0.01) (Fig. 2c). Thus inhibition of G1/S transition was accompanied by cyclin E1 downregulation.
Cyclin D1 regulates entry into and progression through G1 in response to the extracellular environment [22] by inactivating Rb, thus freeing E2F for cyclin E1 transcriptional activation. Therefore, we assessed cyclin D1 protein level. Cyclin D1 ran as a 37-kDa doublet, likely representing different splice variants [23]. There was no significant difference in cyclin D1 protein level between culture conditions (Fig. 2b), showing that the decrease in cyclin E1 was not secondary to decreased cyclin D1.
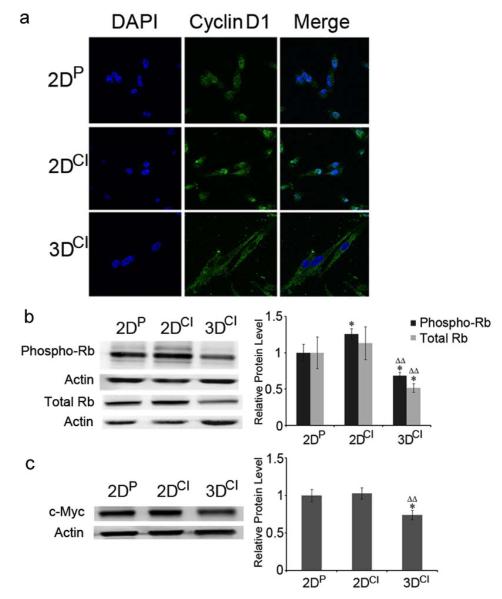
Cyclin D1 is primarily cytoplasmic in 3D collagen culture
Since cyclin D1 is regulated by multiple mechanisms, including changes in subcellular localization [24], its localization was also assessed. Interestingly, cyclin D1 was predominantly nuclear in 2D cultures and predominantly cytoplasmic in 3D cultures (Fig. 3a). Generation of a heat map to show staining intensity (Fig. S1) emphasizes cyclin D1 absence from the nucleus in 3D cultures. Since cyclin D1 exhibits its regulatory function in the nucleus, its presence in the cytoplasm indicated a potential continued activation of Rb and thus transcriptional downregulation of cyclin E1, consistent with the decrease seen in cyclin E1 mRNA. To investigate if cyclin D1 cytoplasmic localization affected phosphorylation and thus activity of Rb, western blotting was performed using an antibody recognizing Rb phosphorylated on Ser780, a cyclin D-cdk4/6 phosphorylation site [25]. Ser780 phosphorylation was decreased in 3D as compared to 2D cultures as was total Rb (Fig. 3b). There was also a significant increase of phospho-Rb in 2D collagen compared to 2D plastic, consistent with increased Ki-67 (Fig. 1d) in 2D collagen cultures.
Fig. 3.
Cyclin D1 relocalized from the nucleus to the cytoplasm in 3D collagen culture. a Confocal images of cyclin D1 in MDA-MB-231 cells. Cyclin D1 staining was visualized with Alexa Fluor 488 conjugated secondary antibody (green). Nuclei were stained with DAPI (blue). Magnification 63X. b Western blots of phospho-Rb (Ser780) and Rb in 2DP, 2DCI and 3DCI MDA-MB-231 cells. *p < 0.05 vs. 2DP, ΔΔp < 0.01 vs. 2DCI. c Western blot of c-Myc in 2DP, 2DCI and 3DCI of MDA-MB-231 cells. *p < 0.05 vs. 2DP, ΔΔp < 0.01 vs. 2DCI. The relative quantity of phospho-Rb, total Rb or c-Myc protein was calculated as in Fig. 2b
Positive regulators of cyclin E1 were decreased in 3D collagen culture
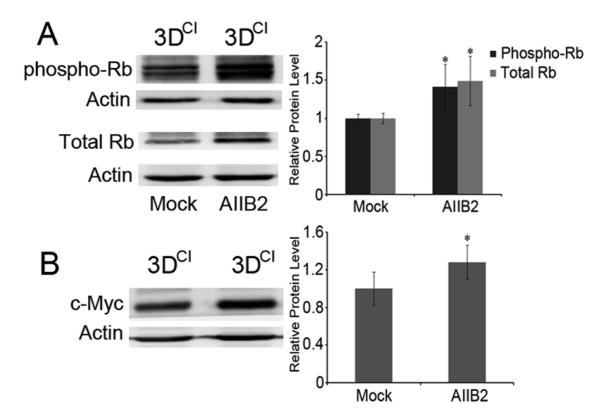
The transcription factor c-Myc is expressed in response to extracellular signals such as mitogens and adhesion and induces expression of cyclin D, cdk4, and E2F, among others. c-Myc was also reported to upregulate cyclin E1 in response to 2D collagen attachment [26]. Therefore, we monitored c-Myc protein level, to determine if it was affected by 3D collagen culture. 3D collagen culture decreased c-Myc by 26% compared to 2D cultures (Fig. 3c) consistent with the increased G1 population, decreased Rb phosphorylation, and decreased cyclin E1 mRNA.
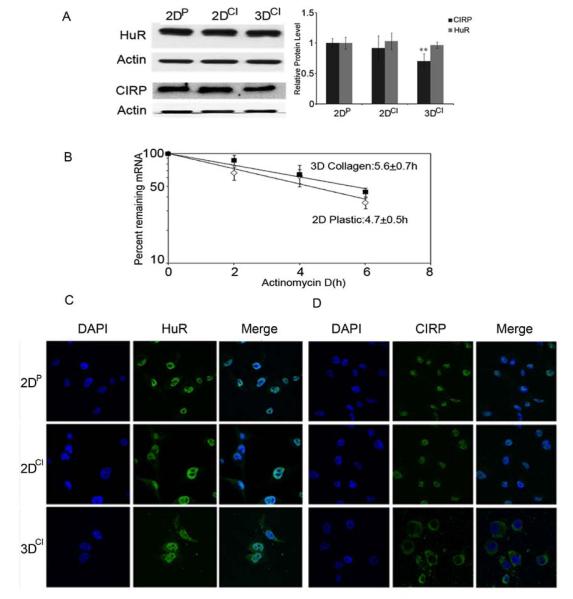
In addition to transcriptional control, cyclin E1 mRNA stability is positively regulated in breast cancer cells by the RNA binding proteins HuR and CIRP [17, 27]. To ask if regulation of these proteins could also underlie changes in cyclin E1, we assessed their relative protein levels. Western analysis showed no significant difference in HuR while CIRP was decreased by 30% in 3D as compared to 2D cultures (Fig. 4a). CIRP and HuR localization was also assessed as they shuttle from nucleus to cytoplasm to stabilize their mRNA targets [28, 29]. While HuR was primarily nuclear in all conditions, CIRP was cytoplasmic in 3D cultures (Fig. 4c and d). Despite this, cyclin E1 mRNA stability was not significantly altered by 3D culture (Fig. 4b) showing that mRNA destabilization did not contribute to the decrease in cyclin E1 mRNA. It’s possible that either the decrease in CIRP or its relocalization to the cytoplasm represses cyclin E1 translation [30] or affects other CIRP target mRNAs to stall G1 phase progression [31].
Fig. 4.
CIRP was decreased and relocalized to the cytoplasm in 3D collagen culture. a Western blots for HuR and CIRP in 2DP, 2DCI and 3DCI MDA-MB-231 cells. ** p < 0.01 vs. 2DP. The relative quantity of HuR and CIRP proteins was calculated as in Fig. 2b. b Half-life determination for cyclin E1 mRNA. Total RNA was extracted from cells grown on 2DP or in 3DCI at the indicated time after addition of actinomycin D. Real time RT-PCR was used to analyze cyclin E1 mRNA level. Data were normalized to GAPDH mRNA or 18s rRNA and plotted on a semi-logarithmic scale. c,d Confocal images of HuR and CIRP localization. HuR or CIRP staining was visualized with Alexa Fluor 488 conjugated secondary antibody (green). Nuclei were stained with DAPI (blue). Magnification 63X
β1 integrin mediated downregulation of cyclin E1
We next asked how 3D collagen could activate signaling pathways that result in downregulation of cyclin E1. Cellular interactions with collagen matrices are mediated by specific receptors, including integrins, with α2β1 integrin being the primary heterodimer on epithelial cells [32]. Western blotting for β1 integrin was performed to see if levels differed in the 3 culture conditions. As shown in Fig. 5a, there was a marked increase of β1 integrin in 3D collagen. We always saw two bands on β1 integrin immunoblots, similar to reports by others and to other cell lines (BD Biosciences, San Jose, CA). As its possible that both bands represent β1 integrin, but with different post-translational modifications, we included both bands for quantitation of β1 integrin level. As α2β1 integrin is a collagen I receptor responsible for delivering ECM signals to the cells [12], and the cells were surrounded by collagen I, increased β1 integrin was likely a functional adaptation to this environment.
Fig. 5.
β1 integrin was dramatically increased by 3D collagen culture. a Western blot for β1 integrin in 2DP, 2DCI and 3DCI MDA-MB-231 cells. **p < 0.01 vs. 2DP, ΔΔp < 0.01 vs. 2DCI. The relative quantity of β1 integrin protein (both bands) was calculated after normalization to actin, expressed as mean ± SD and assessed by Student’s t test. b Western blot for cyclin E1 in 3DCI cultures of MDA-MB-231 cells with (AIIB2) or without AIIB2 (mock) antibody. *p < 0.05. c Confocal images of cyclin D1 in 3DCI MDA-MB-231 cells with or without AIIB2 treatment (63X magnification). d Quantitation of Ki-67 in 3DCI MDA-MB-231 cells with or without AIIB2 treatment. Percentage of Ki-67+ cells was calculated by counting at least 10 microscope fields at 40X (at least 600 cells) for each condition. **p < 0.01
To determine if reduced cyclin E1 expression required β1 integrin interaction with collagen I, a function-blocking antibody (AIIB2) that binds β1 integrin extracellular domain was used to disrupt β1 integrin association with collagen [18, 19]. Cells were cultured in 3D collagen for 24h and then treated with AIIB2 or IgG1 (mock) for 2h. Cyclin E1 increased by 34% after blocking β1 integrin function and cell proliferation increased by 10% compared to the mock group (Fig. 5b and d). Despite the increase in cyclin E1, cyclin D1 remained cytoplasmic upon inhibition of β1 integrin (Fig. 5c), suggesting that cyclin E1 response to blocking β1 integrin function differs from that of cyclin D1. Notably, there was a dramatic morphological change from spindle to rounded cell shape after treatment, indicating that AIIB2 disrupted cell attachment. Cell rounding was also observed in 2D cultures treated with AIIB2 (Fig. S2b), but cell proliferation was significantly decreased (Fig. S2a). These results indicated that inhibition of β1 integrin association with collagen rescued cell proliferation in 3D collagen cultures by upregulating cyclin E1 without changing cyclin D1 localization. Blocking β1 integrin function also restored Rb phosphorylation as well as Rb and c-Myc levels in 3D collagen culture (Fig. 6). The increased phospho-Rb and c-Myc were in agreement with increased cyclin E1 and increased cell proliferation upon inhibition of β1 integrin function. Given the dramatic change in cell shape, it is possible that at least some of these effects resulted from the multiple downstream changes that occur due to the cell shape change.
Fig. 6.
β1 integrin inhibition increased Rb and its phosphorylation as well as c-Myc in 3D collagen. a Western blots of phospho-Rb and total Rb in 3DCI cultures of MDA-MB-231 cells with (AIIB2) or without (Mock) β1 integrin antibody. Cells were grown in 3DCI for 24h and then treated with AIIB2 or IgG1 for 2h. *p < 0.05. b Western blot of c-Myc in 3DCI cultures of MDA-MB-231 cells with or without AIIB2 antibody. *p < 0.05. The relative quantity of phospho-Rb, total Rb or c-Myc was calculated as in Fig. 2b
In order to determine if a 3D collagen I environment regulates the cell cycle of other breast cancer cell lines as it does that of MDA-MB-231 cells, we assessed effects on MCF-7, SKBR3, and Hs578T breast cancer cell lines. MCF-7 and SKBR-3 cells were both derived from pleural effusions of adenocarcinomas and are luminal subtype, while Hs578T cells were derived from a patient carcinosarcoma and similar to MDA-MB-231 cells, are basal B subtype and invasive. Fig. S3 shows that of these cell lines, only MCF-7 cells downregulated cyclin E1 in response to 3D collagen culture, as compared to culture on 2D plastic. SKBR3 cells showed minimal change while Hs578T cells increased both cyclin E1 and cyclin D1. Interestingly, β1 integrin decreased in 3D collagen in the non-invasive MCF-7 and SKBR3 cells, but increased dramatically in invasive Hs578T cells. Phospho- and total Rb decreased in all 3 cell lines in 3D culture (Fig. S3), and cyclin D1 was again primarily cytoplasmic in all 3D cultures (Fig S4). These results show that cyclin E1 regulation by 3D collagen culture differs amongst diverse breast cancer cell lines, but regulation of cyclin D1 appears to be similar.
Discussion
We have shown that a 3D environment composed of collagen I suppresses proliferation of a highly aggressive human breast cancer cell line. The suppression of proliferation resulted from a delay in S phase entry from G1 and an increase in G0 cells. Both cyclin E1 protein and mRNA were decreased, consistent with the inhibition of G1/S transition, while cyclin D1 protein level was not affected. There was a dramatic shift in cyclin D1 localization from primarily nuclear to cytoplasmic in 3D collagen, decreased phosphorylation of Rb, a decrease in total Rb and c-Myc proteins, and a dramatic increase in β1 integrin, a component of the collagen I receptor in epithelial cells. Inhibition of β1 integrin function in 3D collagen increased cell proliferation and expression of cyclin E1, Rb, phospho-Rb, and c-Myc, while cyclin D1 remained cytoplasmic. These results show that cyclin E1 can be negatively regulated by the ECM downstream of β1 integrin, bypassing cyclin D1.
In contrast to the results with the highly invasive MDA-MB-231 cells, three other breast cancer cell lines responded disparately to 3D collagen culture. Invasive Hs578T cells upregulated β1 integrin but also increased cyclins E1 and D1. Like MDA-MB-231 cells, non invasive MCF-7 cells downregulated cyclin E1, but unlike them, downregulated β1 integrin. This downregulation of β1 integrin (also seen in SKBR3 cells) is similar to what would be expected for normal ME cells cultured in 3D collagen as β1 integrin has been implicated in positive regulation of ME cell proliferation both in vitro and in vivo [33], and is necessary for formation of mammary tumors in murine models [34]. Disruption of β1 integrin function results in decreased ME cell proliferation in a transgenic mouse model [35] and in both 2D and 3D tissue culture models [36, 37]. Proliferation of an ME cell line on 2D collagen was positively regulated via β1 integrin induction of c-Myc through activation of the Src and MAPK signaling pathways [38]. c-Myc upregulated cyclin E1 expression, downregulated the cyclin-dependent kinase inhibitor (CKI) p27, and increased phosphorylation of Rb by cyclin E/cdk2 complexes [26]. Similarly, in normal primary human ME cells, α2β1 integrin interaction with a 2D collagen matrix stimulated expression of cyclin E1 and cdk2 as well as cyclin E1/cdk2 complex formation [12]. Consistent with these studies, we noted that 2D collagen culture also increased proliferation of MDA-MB-231 breast cancer cells, while blocking β1 integrin reduced proliferation. Likewise, when MDA-MB-231 cells were cultured in 3D Matrigel, a reconstituted basement membrane composed mainly of collagen IV and laminin, the addition of inhibitory β1-integrin antibody reduced proliferation [19] and reverted the malignant phenotype of HMT3522-T4-2 breast cancer cells [39]. In contrast, nontumorigenic ME cells that form acini-like structures in Matrigel, were resistant to β1 integrin inhibition [19]. Neither inhibiting apoptosis nor enhancing proliferation blocks lumen formation in these epithelial structures, indicating that nontumorigenic ME cells and breast cancer cells exert different mechanisms to maintain their phenotype in the same environment [40].
In contrast to 3D Matrigel, we show that 3D collagen culture inhibits proliferation of MDA-MB-231 cells and blocking β1 integrin function rescues this inhibition. Similar to the above studies, the mechanism is at least partly via direct of cyclin E1, as β1 integrin inhibition restores cyclin E1 expression as well as expression of c-Myc and phosphorylation of Rb without influencing cyclin D1 level or cytoplasmic localization. Thus, our results also implicate c-Myc in transcriptional regulation of cyclin E1 and cyclin E1/cdk2 in phosphorylation of Rb in this cell line. Results similar to ours were reported for arterial smooth muscle cells, which arrest in G1 on the surface of polymerized collagen I and proliferate on monomer collagen [10]. In this study, the CKI p27 was induced and suppressed cyclin E1/cdk2 without changing cyclin E1 level. Although cyclin E1 was not studied, 3D collagen likewise arrested glomerular mesangial cells at G0/G1 phase [42], associated with downregulation of E2F and its targets, including c-Myc, and dephosphorylation of Rb. Our findings add to these by showing that β1 integrin interaction with collagen I can lead to negative regulation of cyclin E1, c-Myc and Rb. We did not address the role of CKIs such as p27. p27 is expressed at differing levels in these cell lines [43], thus we cannot rule out a possible role in inhibiting cyclin E/cdk2 and/or other cyclin/cdk complexes in 3D collagen, contributing to decreased proliferation.
To our knowledge, this is the first study to show cyclin D1 cytoplasmic localization in response to 3D collagen. This cytoplasmic localization was consistently seen in the four cell lines studied. Cyclin D1 overexpression in cancer is often caused by disruption of its degradation [44], which has been shown to accelerate mammary carcinogenesis. At the G1/S boundary, cyclin D1 is phosphorylated by GSK3β followed by nuclear export to the cytoplasm, where cyclin D1 is recognized by SCFFbx4/αB-crystallin ligase, ubiquitinated, and targeted to the 26S proteasome. Knockdown of either Fbx4 or αB-crystallin inhibits cyclin D1 proteolysis and leads to its nuclear accumulation and cell transformation [45]. MDA-MB-231 cells lack αB-crystallin due to a chromosomal deletion [46]. In light of this, the observation that in 3D collagen, cyclin D1 changed from being primarily nuclear to cytoplasmic and remained there after integrin inhibition in MDA-MB-231 cells, was surprising. Our results suggest that cyclin D1 may be phosphorylated by GSK3β in response to a 3D collagen environment, resulting in its inactivation by nuclear export. The ability to directly regulate cyclin E1 in the absence of nuclear cyclin D1 may be an adaptation of MDA-MB-231 and other breast cancer cells to regulate survival and growth in a primarily collagen environment after invasion and during metastasis.
We had shown previously that the RNA binding proteins HuR and CIRP stabilize cyclin E1 mRNA, contributing to its overexpression in breast cancer cells. As microenvironment affects mRNA decay [47], we asked if these proteins decreased in 3D collagen cultures. HuR did not change but CIRP decreased and relocalized to the cytoplasm upon 3D collagen culture. Despite this, cyclin E1 mRNA half-life was not changed, suggesting that if the changes in CIRP contribute to cyclin E1 downregulation it is via another mechanism [30], or effects on other targets.
In summary, our studies show that a 3D environment composed of collagen I can control the cell cycle in part through regulation of cyclin E1. In highly invasive MDA-MB-231 cells this regulation is through β1 integrin-activated signaling pathways that decrease c-Myc and ultimately result in downregulation of cyclin E1. The downstream signaling pathways as well as the control of β1 integrin function in differentially affecting proliferation need to be further investigated, as do the inherent differences in breast cancers.
Supplementary Material
Supplementary Data
Acknowledgments
This work was supported by the National Cancer Institute-National Institutes of Health R01 CA095898 to RSH. We thank Therese Mitchell and Tamara Howard for technical support. AIIB2 monoclonal antibody (Dr. Caroline H. Damsky) was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. Images were generated in the UNM Cancer Center Microscopy Facility, supported from NCRR 1 S10 RR14668, NSF MCB9982161, NCRR P20 RR11830, NCI P30 CA118100, NCRR S10 RR19287, NCRR S10 RR016918, the UNM HSC, and the UNM Cancer Center.
References
- 1.Duffy MJ, Crown J. A personalized approach to cancer treatment: how biomarkers can help. Clinical chemistry. 2008;54:1770–1779. doi: 10.1373/clinchem.2008.110056. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Steeg PS, Price JE, Krishnamurthy S, Mani SA, Reuben J, Cristofanilli M, Dontu G, Bidaut L, Valero V, Hortobagyi GN, Yu D. Breast cancer metastasis: challenges and opportunities. Cancer research. 2009;69:4951–4953. doi: 10.1158/0008-5472.CAN-09-0099. [DOI] [PubMed] [Google Scholar]
- 3.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes & development. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 5.Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: the potential roles of c-Myc, cyclin D1 and cyclin E. Endocr Relat Cancer. 2005;12(Suppl 1):S47–59. doi: 10.1677/erc.1.00993. doi: 12/Supplement_1/S47 [pii] 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- 6.Keyomarsi K, O’Leary N, Molnar G, Lees E, Fingert HJ, Pardee AB. Cyclin E, a potential prognostic marker for breast cancer. Cancer Res. 1994;54:380–385. [PubMed] [Google Scholar]
- 7.Sieuwerts AM, Look MP, Meijer-vanGelder ME, Timmermans M, Trapman AM, Garcia RR, Arnold M, Goedheer AJ, de Weerd V, Portengen H, Klijn JG, Foekens JA. Which cyclin E prevails as prognostic marker for breast cancer? Results from a retrospective study involving 635 lymph node-negative breast cancer patients. Clin Cancer Res. 2006;12:3319–3328. doi: 10.1158/1078-0432.CCR-06-0225. doi: 12/11/3319 [pii] 10.1158/1078-0432.CCR-06-0225. [DOI] [PubMed] [Google Scholar]
- 8.Potemski P, Kusinska R, Pasz-Walczak G, Piekarski JH, Watala C, Pluciennik E, Bednarek AK, Kordek R. Prognostic relevance of cyclin E expression in operable breast cancer. Med Sci Monit. 2009;15:MT34–40. doi: 869544 [pii] [PubMed] [Google Scholar]
- 9.Henriet P, Zhong ZD, Brooks PC, Weinberg KI, DeClerck YA. Contact with fibrillar collagen inhibits melanoma cell proliferation by up-regulating p27KIP1. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10026–10031. doi: 10.1073/pnas.170290997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koyama H, Raines EW, Bornfeldt KE, Roberts JM, Ross R. Fibrillar collagen inhibits arterial smooth muscle proliferation through regulation of Cdk2 inhibitors. Cell. 1996;87:1069–1078. doi: 10.1016/s0092-8674(00)81801-2. [DOI] [PubMed] [Google Scholar]
- 11.Cho MK, Suh SH, Lee CH, Kim SG. Bovine type I collagen inhibits Raw264.7 cell proliferation through phosphoinositide 3-kinase- and mitogen-activated protein kinase-dependent down-regulation of cyclins D1, A and B1. Biochim Biophys Acta. 2005;1744:47–57. doi: 10.1016/j.bbamcr.2004.11.004. doi: S0167-4889(04)00310-6 [pii] 10.1016/j.bbamcr.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Klekotka PA, Santoro SA, Ho A, Dowdy SF, Zutter MM. Mammary epithelial cell-cycle progression via the alpha(2)beta(1) integrin: unique and synergistic roles of the alpha(2) cytoplasmic domain. The American journal of pathology. 2001;159:983–992. doi: 10.1016/s0002-9440(10)61774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudjonsson T, Ronnov-Jessen L, Villadsen R, Bissell MJ, Petersen OW. To create the correct microenvironment: three-dimensional heterotypic collagen assays for human breast epithelial morphogenesis and neoplasia. Methods (San Diego, Calif.) 2003;30:247–255. doi: 10.1016/s1046-2023(03)00031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronnov-Jessen L, Bissell MJ. Breast cancer by proxy: can the microenvironment be both the cause and consequence? Trends in molecular medicine. 2009;15:5–13. doi: 10.1016/j.molmed.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsdale T, Bard J. Collagen substrata for studies on cell behavior. The Journal of cell biology. 1972;54:626–637. doi: 10.1083/jcb.54.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wozniak MA, Keely PJ. Use of three-dimensional collagen gels to study mechanotransduction in T47D breast epithelial cells. Biological Procedures Online. 2005;7:144–161. doi: 10.1251/bpo112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo X, Hartley RS. HuR contributes to cyclin E1 deregulation in MCF-7 breast cancer cells. Cancer research. 2006;66:7948–7956. doi: 10.1158/0008-5472.CAN-05-4362. [DOI] [PubMed] [Google Scholar]
- 18.Hall DE, Reichardt LF, Crowley E, Holley B, Moezzi H, Sonnenberg A, Damsky CH. The alpha 1/beta 1 and alpha 6/beta 1 integrin heterodimers mediate cell attachment to distinct sites on laminin. The Journal of cell biology. 1990;110:2175–2184. doi: 10.1083/jcb.110.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park CC, Zhang H, Pallavicini M, Gray JW, Baehner F, Park CJ, Bissell MJ. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer research. 2006;66:1526–1535. doi: 10.1158/0008-5472.CAN-05-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porter DC, Keyomarsi K. Novel splice variants of cyclin E with altered substrate specificity. Nucleic acids research. 2000;28:E101. doi: 10.1093/nar/28.23.e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwell RM, Porter DC, Danes C, Keyomarsi K. Processing of cyclin E differs between normal and tumor breast cells. Cancer research. 2000;60:481–489. [PubMed] [Google Scholar]
- 22.Diehl JA. Cycling to cancer with cyclin D1. Cancer biology & therapy. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 23.Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z, Senderowicz AM, Conti CJ, Knudsen ES. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. The Journal of biological chemistry. 2003;278:30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 24.Gladden AB, Diehl JA. Location, location, location: the role of cyclin D1 nuclear localization in cancer. Journal of cellular biochemistry. 2005;96:906–913. doi: 10.1002/jcb.20613. [DOI] [PubMed] [Google Scholar]
- 25.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Molecular and cellular biology. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benaud CM, Dickson RB. Adhesion-regulated G1 cell cycle arrest in epithelial cells requires the downregulation of c-Myc. Oncogene. 2001;20:4554–4567. doi: 10.1038/sj.onc.1204609. [DOI] [PubMed] [Google Scholar]
- 27.Guo X, Wu Y, Hartley RS. Cold-inducible RNA-binding protein contributes to human antigen R and cyclin E1 deregulation in breast cancer. Molecular carcinogenesis. 2009 doi: 10.1002/mc.20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoki K, Matsumoto K, Tsujimoto M. Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs. The Journal of biological chemistry. 2003;278:48491–48497. doi: 10.1074/jbc.M308328200. [DOI] [PubMed] [Google Scholar]
- 29.Brennan CM, Steitz JA. HuR and mRNA stability. Cellular and molecular life sciences : CMLS. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Experimental cell research. 2007;313:4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. The Journal of cell biology. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCall-Culbreath KD, Zutter MM. Collagen receptor integrins: rising to the challenge. Current Drug Targets. 2008;9:139–149. doi: 10.2174/138945008783502494. [DOI] [PubMed] [Google Scholar]
- 33.Taddei I, Faraldo MM, Teuliere J, Deugnier MA, Thiery JP, Glukhova MA. Integrins in mammary gland development and differentiation of mammary epithelium. J Mammary Gland Biol Neoplasia. 2003;8:383–394. doi: 10.1023/B:JOMG.0000017426.74915.b9. doi: 10.1023/B:JOMG.0000017426.74915.b9 481709 [pii] [DOI] [PubMed] [Google Scholar]
- 34.White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 2004;6:159–170. doi: 10.1016/j.ccr.2004.06.025. doi: 10.1016/j.ccr.2004.06.025 S1535610804002077 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Faraldo MM, Deugnier MA, Lukashev M, Thiery JP, Glukhova MA. Perturbation of beta1-integrin function alters the development of murine mammary gland. EMBO J. 1998;17:2139–2147. doi: 10.1093/emboj/17.8.2139. doi: 10.1093/emboj/17.8.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci U S A. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasqualini R, Hemler ME. Contrasting roles for integrin beta 1 and beta 5 cytoplasmic domains in subcellular localization, cell proliferation, and cell migration. J Cell Biol. 1994;125:447–460. doi: 10.1083/jcb.125.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benaud CM, Dickson RB. Regulation of the expression of c-Myc by beta1 integrins in epithelial cells. Oncogene. 2001;20:759–768. doi: 10.1038/sj.onc.1204152. [DOI] [PubMed] [Google Scholar]
- 39.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. The Journal of cell biology. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. doi: S0092867402010012 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. doi: 10.1083/jcb.200305010 jcb.200305010 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuboi N, Yoshida H, Kawamura T, Furukawa Y, Hosoya T, Yamada H. Three-dimensional matrix suppresses E2F-controlled gene expression in glomerular mesangial cells. Kidney international. 2000;57:1581–1589. doi: 10.1046/j.1523-1755.2000.00002.x. [DOI] [PubMed] [Google Scholar]
- 43.Sweeney KJ, Swarbrick A, Sutherland RL, Musgrove EA. Lack of relationship between CDK activity and G1 cyclin expression in breast cancer cells. Oncogene. 1998;16:2865–2878. doi: 10.1038/sj.onc.1201814. doi: 10.1038/sj.onc.1201814. [DOI] [PubMed] [Google Scholar]
- 44.Pontano LL, Diehl JA. Speeding through cell cycle roadblocks: Nuclear cyclin D1-dependent kinase and neoplastic transformation. Cell division. 2008;3:12. doi: 10.1186/1747-1028-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, Lu F, Rustgi AK, Diehl JA. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin DI, Lessie MD, Gladden AB, Bassing CH, Wagner KU, Diehl JA. Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis. Oncogene. 2008;27:1231–1242. doi: 10.1038/sj.onc.1210738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross J. mRNA stability in mammalian cells. Microbiological reviews. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data