Unexpected Roles for Core Promoter Recognition Factors in Cell-type Specific Transcription and Gene Regulation (original) (raw)
. Author manuscript; available in PMC: 2011 Feb 1.
Published in final edited form as: Nat Rev Genet. 2010 Jul 13;11(8):549–558. doi: 10.1038/nrg2847
Abstract
Until recently, the eukaryotic core promoter recognition complex was generally thought to play an essential but passive role in the regulation of gene expression. However, recent evidence indicates that core-promoter recognition complexes in conjunction with “non-prototypic” subunits may play a critical regulatory role in driving cell specific programs of transcription during development. Furthermore, new roles for components of these complexes have been identified beyond development, for example in mediating interactions with chromatin and in maintaining active gene expression across cell divisions.
Introduction
Until recently it was thought that a universal and highly conserved RNA polymerase II (Pol II) core promoter recognition apparatus initiated transcription in all eukaryotic cells1. Central components of the prototypic preinitiation complex such as TFIID – a complex of TBP (TATA binding protein) and TAFs (TBP associated factors) – were generally considered essential but passive partners that were destined to follow the regulatory instructions provided by sequence specific activators and repressors2. This view came in part from studying a limited set of cell types – for example, yeast, Drosophila S2 cells, and human HeLa cells – which divide rapidly and were preferred for practical reasons such as large-scale production for biochemical analysis or ease of genetic manipulation. In the few cases where more differentiated cell types and tissues had been used, they often comprised a mixture of cell types (e.g. whole rat liver, calf thymus, drosophila embryos)3–5. Furthermore, many experiments in the transcription field have involved using recombinant model genes and promoters and artificial regulators6–10.
More recent studies have shifted toward an analysis of endogenous genes and physiologically relevant regulators observed in the context of nearly homogeneous populations of a single, specific, differentiated cell type and in distinct cell cycle stages. These studies have revealed the requirement for a number of ‘non-prototypic’ core promoter recognition factors, including cell-type specific TAFs and TBP related factors (TRFs). Furthermore, new functions of the prototypical core promoter recognition machinery have been identified. Here we review these studies, which have implications for understanding gene regulation during both somatic and germ cell development – where these factors are increasingly being shown to regulate specific sets of genes – and reveal unexpected functions for core promoter recognition factors more generally in transcriptional regulation and the maintenance of gene expression states.
Prototypical core promoter recognition
Core promoter recognition is the first step in the general mechanism of transcription initiation1. The primary prototypical core promoter recognition factor for eukaryotic mRNA genes is the general transcription factor TFIID, which binds multiple core promoter elements to begin the process of forming preinitiation complexes containing Pol II (Figure 1). RNA polymerase II core promoters in higher eukaryotes are highly diverse and the core promoters of many genes do not contain any of the known core promoter elements. The most recognizable core promoter element is the TATA box, but TATA-containing promoters are actually a minority compared with the aggregate of TATA-less promoters11. It now seems unlikely that one can simply classify promoters into TATA-containing versus TATA-less as there appear to be many potentially diverse TATA-less classes of promoters. However, the prevailing evidence suggests that subunits of the TFIID complex function at most, if not all RNA polymerase II promoters in higher eukaryotes.
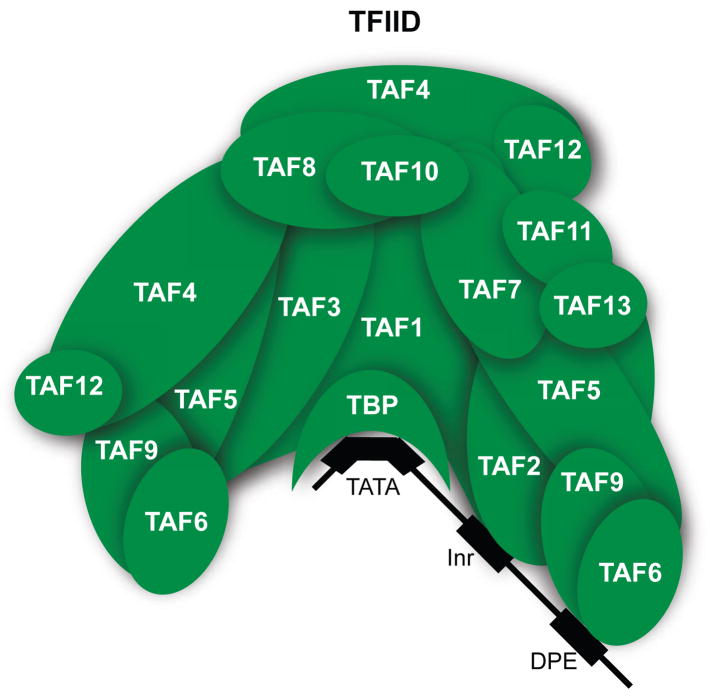
Figure 1. Core promoter recognition by TFIID.
Multiple subunits of the TFIID (transcription factor IID) complex bind core promoter elements12. TBP (TATA binding protein) binds TATA boxes. TAF1 (TBP associated factor 1) and TAF2 (TBP associated factor 2) bind the initiator element (Inr). TAF6 (TBP associated factor 6) and TAF9 (TBP associated factor 9) bind the downstream promoter element (DPE).
TFIID, which consists of TBP and 13-14 TAF subunits, binds core promoter DNA via multiple subunits (e.g. TBP, TAF1, TAF2, TAF6, and TAF9)11 (Figure 1). The TBP subunit of TFIID binds TATA boxes, which when present in promoters, are centered approximately 27 basepairs upstream of the transcription start site12. Several of the TAFs also bind promoter elements downstream of the TATA box. TAF1 and TAF2 bind the initiator, which spans the transcription start site, TAF6 and TAF9 bind the DPE (downstream promoter element), and TAF1 is in close proximity to the DCE (downstream core element) when TFIID is bound to promoters12. Some of the TAF subunits are also the targets of transcriptional activators2,13, allowing TFIID to integrate signals from activators to the core promoter.
The general transcription factor TFIIA aids TFIID in binding core promoters, after which the remaining general transcription machinery can associate, including TFIIB, Pol II, TFIIF, TFIIE, and TFIIH, as well as the mediator coactivator complex. Once formed, preinitiation complexes are competent to initiate transcription. In cells, as opposed to in vitro, this process is far more complicated, primarily due to the DNA being associated with nucleosomes as part of chromatin plus the additional requirement for transcriptional activators, coactivators, chromatin modifying factors, and transcription elongation factors. However, while these additional factors are critical for transcription in cells, they are not considered to be core promoter recognition factors. Recently, subunits of TFIID have been found to display activities that were not expected based on the dogmatic view of core promoter recognition and preinitiation complex formation. Proteins related in sequence to TBP and several TAFs have been discovered that appear to display unique functions during development, differentiation, and cell proliferation. These proteins and the functions discussed below are summarized in Table 1.
Table 1.
Non-prototypic functions of components of core promoter recognition complexes.
| Factor | System studied | Functions discussed | References |
|---|---|---|---|
| TBP | Human HeLa cells | Mitotic bookmarking | 64,65,67 |
| TRF2 | C. elegans, Xenopus laevis | Early embryogenesis | 22–24 |
| Drosophila melanogaster | Metamorphosis | 26 | |
| Drosophila melanogaster S2 cells | Transcription (e.g. Histone H1) | 53,54 | |
| Mouse | Spermatogenesis | 14–16,18–21 | |
| TRF3 | Xenopus laevis | Gastrulation | 24,29,30 |
| Zebrafish | 28 | ||
| Mouse embryonic stem cells | Embryogenesis, hematopoiesis | 27,45,46 | |
| Mouse C2C12 cells | Hematopoiesis | 47,48 | |
| Mouse | Myogenesis | 30–32 | |
| Oogenesis | |||
| TAF3 | Zebrafish | Hematopoiesis | 46 |
| Mouse embryonic stem cells | Hematopoiesis | 46 | |
| Mouse C2C12 cells | 47,48 | ||
| Human U2OS cells | Myogenesis | 59 | |
| Anchors TFIID to H3K4me3 | |||
| TAF4b | Mouse | Oogenesis, spermatogenesis | 36–42 |
| TAF7L | Mouse | Spermatogenesis | 43,44 |
| TAF8 | Mouse | Adipogenesis | 50 |
| No hitter (TAF4 homolog) | Drosophila melanogaster | Spermatogenesis | 34,35 |
| Cannonball (TAF5 homolog) | Drosophila melanogaster | Spermatogenesis | 33,35 |
| Meiosis I arrest (TAF6 homolog) | Drosophila melanogaster | Spermatogenesis | 34,35 |
| Spermatocyte arrest (TAF8 homolog) | Drosophila melanogaster | Spermatogenesis | 34,35 |
| Ryan express (TAF12 homolog) | Drosophila melanogaster | Spermatogenesis | 34,35 |
Germ Cell Differentiation
TRF2 functions in germ cell differentiation
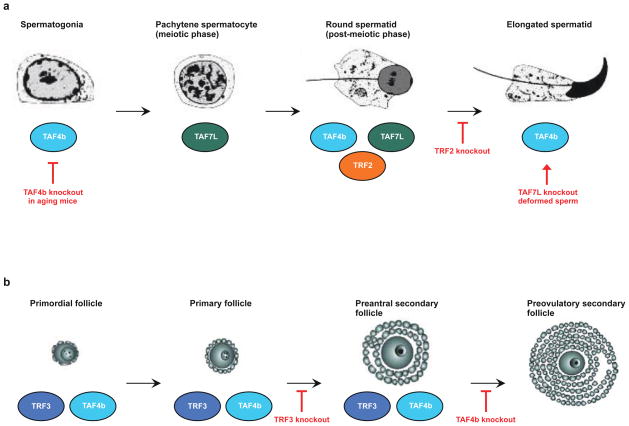
TRF2 (also known as TLF, TLP, TRP, or TBPL1) was independently discovered by sequence similarity to TBP in organisms ranging from C. elegans to human14–18. Although highly similar in sequence to TBP, TRF2 does not bind the TATA box14–16,18. In mammals, TRF2 is most highly expressed in testes, although it is also found at lower levels in other tissues14–16,18, and a primary defect of TRF2 knockout in mice is a deficiency in spermatogenesis (Figure 2A)19,20. Expression of TRF2 is tightly controlled in developing spermatocytes, with high levels of expression first occurring in step 6–7 spermatids20. In TRF2 knockout mice, defects in spermatogenesis first appear at step 7, differentiation to elongated spermatids does not occur and transcription of multiple postmeotic, testes-specific genes is severely decreased19,20. Further investigation showed that early stage spermatids in these mice show a defect in formation of the chromocenter, which is a structure containing condensed centromeric heterochromatin that is involved in chromatin organization21. It is not yet clear whether the deficiency of spermatogenesis in the TRF2 knockout is due entirely to altered gene expression and chromocenter formation.
Figure 2. Roles of TRFs and TAFs in germ cell differentiation.
(a) Overview of mouse spermatogenesis68. TRF2 (TBP related factor 2), TAF4b (TBP associated factor 4b), and TAF7L (TBP associated factor 7L) are expressed where indicated and the effect of knocking out each of these factors is shown19–21,42–44. (b) Overview of a subset of the stages of mouse oogenesis31. TRF3 and TAF4b are thought to be expressed where indicated and the effect of knocking out each of these factors is shown31,32,37,38. In the case of TAF4b, knockout could affect multiple stages; the last of these stages is indicated38.
In addition to functioning in germ cell differentiation, TRF2 has been found to function in embryogenesis in some organisms. For example, in C. elegans and Xenopus, TRF2 is required for early embryogenesis and for the proper transcription of many genes22–24. TRF2 functions both during germ cell differentiation and during the onset of metamorphosis in Drosophila, where mutations in TRF2 result in alterations in the levels and temporal delays in transcription of genes normally regulated by ecdysone during early metamorphosis25,26. Indeed, it seems likely that as more cases are studied, although germ-cell development may initially emerge as a primary process regulated by atypical TRFs and TAFs, the specific functions of these factors may turn out to be utilized in diverse contexts in different organisms.
TRF3 functions in germ cell differentiation
TRF3 (also known as TBP2) was first identified during a search of the initial draft of the human genome for predicted proteins with sequence similarity to human TBP27. The similarity between TRF3 and TBP is limited to their C-terminal DNA binding domains, which are nearly identical; by contrast, the N-terminal domains share little sequence similarity. Genes encoding TRF3 have also been found in the mouse, frog, and zebrafish genomes, but not those of Drosophila and C. elegans27–29. TRF3 was characterized biochemically and found to bind TATA boxes, to interact with TFIIA and TFIIB, and to direct basal transcription, properties it shares with TBP28,29. Hence, TRF3 can be thought of as a replacement for TBP, with respect to these activities.
In early studies TRF3 protein was shown to be expressed at various levels in many human and mouse tissues and cell lines27. However, other groups have found TRF3 mRNA to be most highly expressed in the testis and ovary of zebrafish and Xenopus, and in oocytes in mice28,30. The reason for these different observed expression levels and patterns is not known; although the use of different antibodies and extraction methods may account for this in part, at the current time the distinct expression patterns observed in different laboratories remain unresolved.
In mice, it is clear that TRF3 is highly expressed in the ovary27,30,31. Specifically, the TRF3 protein is found in the nuclei of mouse oocytes during folliculogenesis and levels decrease during ovulation. Interestingly, TBP was not detected in oocytes during the stages of folliculogenesis in which TRF3 was found in the nucleus31. TRF3 knockout are viable, with the only apparent phenotype being female sterility resulting from disrupted oocyte growth and follicular development32. Pol II activity and levels of H3K4me3 were significantly decreased in these animals and the expression of hundreds of genes was altered. A significant number of the most strongly down-regulated genes are oocyte specific, indicating that TRF3 is required for normal levels of transcription in developing oocytes. ChIP assays confirmed that TRF3 occupies the promoters of a number of these genes in developing oocytes in wild type mice (Figure 2B), suggesting that it effectively replaces TBP in directing transcription during folliculogenesis.
Non-prototypic TAFs in germ cell differentiation
A number of non-prototypic TAFs also have roles in germ cell differentiation. The protein encoded by the Drosophila cannonball gene, which is a homolog of Drosophila TAF5, is selectively expressed in primary spermatocytes and is required to establish proper levels of transcription of multiple genes involved in spermatid differentiation33. An additional four alternative TAFs are expressed in primary Drosophila spermatocytes: no hitter, a homolog of TAF4; meiosis I arrest, a homolog of TAF6; spermatocyte arrest, a homolog of TAF8; and ryan express, a homolog of TAF1234. All four of these testis-specific TAFs are required for proper spermatocyte differentiation and control expression of a common set of genes involved in the differentiation process. Further investigation showed that testis-specific TAFs are localized at the promoters of genes involved in spermatid development35. The presence of wild type testis-specific TAFs at the target promoters in these studies correlated with reduced occupancy of polycomb repressive complex 1 (PRC1), a chromatin modifying complex that mediates transcriptional silencing, and increased H3K4me3, a mark for active transcription. Together these observations suggest that Drosophila testis-specific TAFs function during transcriptional activation of developmental genes in primary spermatocytes, likely by reducing levels of PRC1 found at the promoter. Exactly how the TAFs reduce PRC1 levels is not fully understood. However, much of the pool of testis-specific TAFs localizes with PRC1 in a sub-compartment of spermatocyte nucleoli, and this localization requires the testis-specific TAFs35. A portion of the testis-specific TAFs might function to relocalize PRC1 away from genes required for spermatid development, thereby de-repressing their transcription.
TAF4b was initially discovered as a cell-type specific TAF, uniquely found in a human B cell line36. However, when its expression was analyzed in mouse tissues, it was found to be most highly expressed in the testes and ovary37. In the mouse ovary, TAF4b mRNA is uniquely localized to the granulosa cells of the ovarian follicle, and knockout of TAF4b resulted in infertile females with smaller ovaries that lack mature follicles (Figure 2B)37. Further studies showed that TAF4b promotes granulosa cell proliferation and is required for the survival of these cells38. TAF4b knockout mice have reduced expression of many ovarian specific genes, and the overall gene expression program in ovaries from young knockout mice appear similar to those of aged wild type mice, consistent with an observed acceleration in ovary aging in the knockout mice37,39. Over-expression of TAF4b in a rat spontaneously immortalized granulosa cell line significantly increased the expression of a range of genes, notably including c-jun40; this increase seems to be cell type specific since it was not observed in NIH/3T3 cells over-expressing TAF4b. ChIP assays showed that TAF4b and c-Jun co-localized to the promoters of several genes, including that of c-jun itself. Moreover, TFIID complexes containing TAF4b have higher transcriptional activity on the c-jun promoter in a reconstituted transcription system than is achieved with the prototypical TFIID complex41. Together these studies indicate that TAF4b works with a specific transcriptional activator, c-Jun, to control transcription of genes involved in follicle growth.
TAF4b is also involved in spermatogenesis in the mouse, where it is expressed in spermatids in adults (Figure 2A)42. Male TAF4b knockout mice are initially fertile, but become infertile as they age; the testes degenerate and germ cells are lost. Furthermore, multiple genes involved in spermatogenesis are expressed at lower levels in TAF4b knockout mice. It will be interesting to determine if TAF4b also interacts with a specific transcriptional activator to control spermatogenesis.
Finally, a paralog of TAF7, TAF7L, is also involved in male germ cell development in mice43. TAF7L is expressed in male germ cells throughout differentiation and is found in the nucleus of spermatids (Figure 2A). As germ cell development progresses, TAF7L expression increases, which correlates with increased TBP expression and decreased TAF7 expression. In spermatocytes, TAF7L is associated with TBP, whereas TAF7 is not. Knockout of the TAF7L gene in mice leads to the development of deformed sperm, although the mice are fertile44. Gene expression profiling revealed six transcripts that were decreased in abundance in testis from the knockout mice, although their relevance to spermatogenesis was not studied.
Somatic cell differentiation
How are transcriptional programs established in the diverse differentiation pathways that occur in multi-cellular organisms and how do these transcriptional programs direct specific differentiation pathways? Research aimed at answering these questions has largely focused on transcriptional activators and repressors, which are known to play critical roles in directing cell-type specific transcription. More recently, however, non-prototypic core promoter recognition factors have also been found important for directing differentiation
TRF3 and TAF3 function in embryogenesis
Decreased TRF3 levels in zebrafish embryos were found to cause defects in mesoderm patterning28. In addition, during Xenopus embryogenesis, TRF3 is required for gastrulation29 for the normal expression of nearly 900 genes in the developing Xenopus embryo24. Thus, a protein considered to be a non-prototypic core promoter recognition factor controls a transcriptional program involving a very large number of genes.
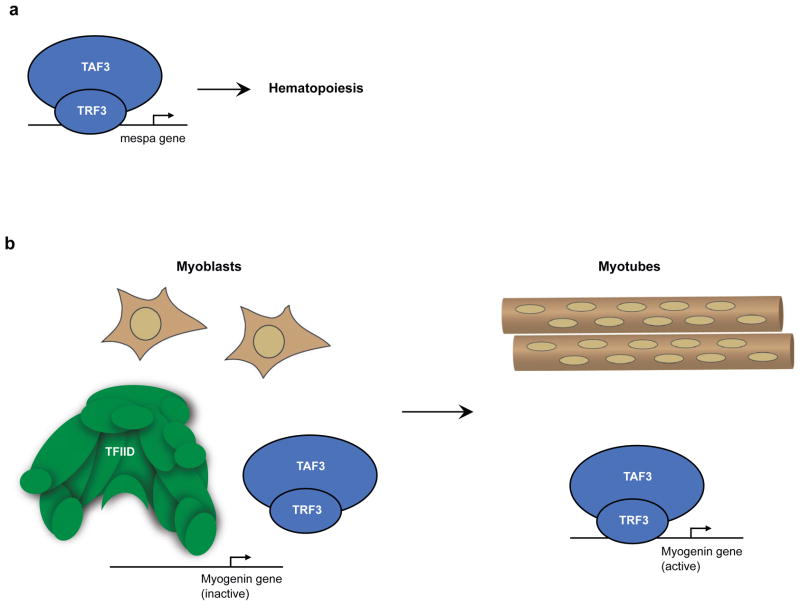
Further studies of TRF3 during zebrafish embryogenesis revealed that it is required for hematopoiesis45 (Figure 3A). Expression profiling and ChIP assays showed that TRF3 was required for the developmentally regulated transcription of the mespa gene, which encodes a basic helix-loop-helix transcription factor that is critical for hematopoiesis45. The mespa promoter was bound by TRF3, but not by TBP. Depletion of mespa resulted in developmental defects that were similar to those observed with depletion of TRF3 and, importantly, ectopic expression of mespa in the TRF3 depleted embryo restored normal development. Ultimately it was shown that the differentiation of mesoderm into the hematopoietic lineage involved binding of TRF3 to the mespa promoter. A more recent study found that TRF3 functions in conjunction with TAF3 to initiate hematopoiesis in the zebrafish embryo46. TAF3 binds TRF3, associates with the mespa promoter, and is required for hematopoiesis. Extension of this analysis to mice showed that TRF3 and TAF3 bind the promoter of the Mesp1 gene, the mouse ortholog of the zebrafish mespa gene, and that TRF3 is also required to initiate hematopoiesis in a mouse ES model system46.
Figure 3. Roles of TRF3 and TAF3 in somatic cell differentiation.
(a) TRF3 (TBP related factor 3) and TAF3 (TBP associated factor 3) function in hematopoiesis. During embryogenesis in zebrafish, TRF3 and TAF3 associate with the core promoter of the mespa gene, which encodes a transcription factor that is critical for hematopoiesis45,46. (b) Proposed TRF3 and TAF3 function in myogenesis. When mouse myoblasts differentiate into myotubes, levels of multiple TFIID subunits decrease47,48. By contrast, TAF3 levels remain constant, as do levels of TRF3. TRF3 and TAF3 form a complex in myotubes and are found together at the promoter of the myogenin gene, which encodes a transcriptional activator that is critical for myogenesis.
There is evidence to suggest that the coupling of TRF3 and TAF3 to facilitate a specific differentiation pathway is not limited to hematopoiesis. In studies of TFIID in skeletal muscle differentiation, the levels of prototypical TFIID subunits (TBP, TAF1, and TAF4) were found to be dramatically reduced when C2C12 myoblasts were induced to differentiate into myotubes (Figure 3B)47. By contrast TAF3, and other components of the general transcription machinery (Pol II, TFII-A, B, E, F, and H) were present at similar levels in the nuclei of myoblasts and myotubes. These observations held true in myoblasts and myofibers isolated from mouse skeletal muscle. Interestingly, the low levels of TRF3 remained relatively constant when C2C12 cells differentiated into myotubes, in which TAF3 and TRF3 were found to interact. Depletion of either TAF3 or TRF3 inhibited the differentiation of C2C12 cells to myotubes and blocked the expression of MyoD and Myogenin, which are transcription factors known to regulate myogenesis.
ChIP assays showed that TAF3 and TRF3 occupy the Myogenin promoter in myotubes, but not in myoblasts. Subsequent studies showed that TAF3 and TRF3 can mediate activation of transcription from the Myogenin promoter by MyoD in a reconstituted transcription system48. The ability of TAF3/TRF3 to direct transcription from the Myogenin promoter was independent of Mediator subunits, which are also depleted in myotubes. Together these results led to a model in which there is a profound reorganization of the Pol II promoter recognition machinery during myogenesis; TFIID subunits are decreased and TAF3/TRF3 directs transcription of genes encoding key myogenic transcriptional regulators47,48.
There is an ongoing debate about significance of these results from cell lines. Knockout of TRF3 in mice did not appear to affect the development of skeletal muscle or blood32 as might have been anticipated given the documented functions of TRF3 in myogenesis in the C2C12 cell model and hematopoiesis in zebrafish, mouse ES cells, and F9 cells45–48. There are multiple possible explanations for these apparently contradictory results. TRF3 could normally function in myogenesis and hematopoiesis in developing mice, but when knocked out another factor could compensate for the loss of TRF3 in myogenesis and hematopoiesis but not in embryogenesis. The compensatory factor, however, would not be able to replace TRF3’s function in the cellular model systems for myogenesis and hematopoiesis, nor in zebrafish hematopoiesis. Additional experiments (e.g. expression of potential compensatory factors in cells or inactivation of potential factors in the TRF3 knockout mice) could test this possibility. With respect to this possibility it is worth noting that in Xenopus oocytes ectopically expressed TBP can replace TRF3 in driving transcription from specific promoters49. Alternatively, muscle and blood phenotypes may only be displayed in the TRF3 knockout mice under very specific conditions. For example, the TRF3 knockout mice could be deficient in wound healing which requires muscle stem cells to differentiate in a timely manner. It is also possible that the commonly used cellular model systems are not reliable models for some aspects of myogenesis and hematopoiesis. In general, it can be difficult to prove that a specific factor is not involved in a particular complex process given the redundancies of living systems. Clearly, additional experiments employing different strategies need to be performed to test the various possibilities regarding the role of TRF3 in mouse myogenesis, hematopoiesis and other developmental pathways.
Unique functions of TAF8 and TAF10 in development
A non-prototypic TAF plays a key role during adipogenesis, although in this case a wholesale rearrangement of the core promoter recognition machinery does not seem to occur. Roeder and colleagues found that levels of many prototypical TFIID TAFs decrease when 3T3-L1 preadipocytes are induced to differentiate into adipocytes, with the exception of the non-prototypic factor TAF850. By contrast, TAF8 was not detected in preadipocytes, but was upregulated during adipogenesis and found associated with TFIID. TAF8 contains a histone fold domain in its N-terminal region that interacts with other TAFs that contain histone folds, which likely facilitates the association of TAF8 with the TFIID complex. Over-expression of the TAF8 histone fold domain blocked the differentiation of 3T3-L1 preadipocytes to adipocytes, perhaps by blocking the association of endogenous TAF8 with TFIID. This effect could be reversed by over-expression of full length TAF850. Importantly, expression of the TAF8 histone fold domain repressed expression of PPARγ and C/EBPα, two regulators of adipogenesis. Together these observations support a model in which TAF8 association with TFIID during the differentiation of preadipocytes to adipocytes stimulates transcription of genes required for adipogenesis, although the mechanism by which this occurs is not understood.
Knockout of TAF10, a well characterized subunit of TFIID, leads to embryonic lethality in mice51. Conditional inactivation of TAF10 in the embryonic liver caused a dramatic reduction in the size of the liver, suggesting that TAF10 is required for liver development52. When TAF10 was conditionally inactivated in the livers of adult mice, the TFIID complex was found to disassemble, although subunits other than TAF10 were still present in liver cells52. Transcript profiling revealed that inactivation of TAF10 in the developing liver affected expression of only 11% of genes, the majority of which are hepatocyte specific, which is the likely cause of the defects observed in liver development upon TAF10 inactivation.
Non-prototypic Functions Beyond Development
Several additional examples of new functions involving core promoter recognition have emerged in the past several years. The common theme that ties the three examples below together is that a single core promoter recognition factor was found to have a new activity. The three factors, which have already been discussed in this review, are TRF2, TAF3, and TBP, the primary prototypical core promoter recognition factor.
Drosophila TRF2 controls transcription of many genes
Tjian and colleagues immunopurified Drosophila TRF2 and found that it associates with NURF, a nucleosome remodeling factor, and DREF, a transcription factor that controls the expression of cell cycle and proliferation genes53. Biochemical and cell-based experiments showed that TRF2 and DREF direct transcription from the proliferating cell nuclear antigen (PCNA) gene, which encodes a protein required for high fidelity DNA replication; the promoter of the PCNA gene contains a DRE (the binding site for DREF). In a later study, Drosophila TRF2 was found to be required for histone H1 transcription and TRF2 was found to occupy the TATA-less histone H1 promoter54. Interestingly, TRF2 did not occupy the promoter of core histone genes that are directly adjacent to the histone H1 gene within the same repeating histone cluster; instead, the promoters of the four core histone genes were occupied by TBP. This is interesting because the amounts of the linker histone H1 relative to the core histones can vary dramatically in cells during development55,56.
ChIP-chip experiments identified over 1000 genes occupied by TRF2 in Drosophila S2 cells, including a cluster of ribosomal protein genes whose co-regulation requires TRF254. Comparison with TBP ChIP-chip data showed that the genes bound by TRF2 and those bound by TBP were largely non-overlapping (80%). Moreover, while the majority of the TBP bound genes contained a TATA box, the TRF2 bound genes were nearly all devoid of a TATA box. Knock down of TRF2 in salivary glands caused severe growth defects, which were most consistent with TRF2 normally functioning to promote cell growth during development. These studies show that TRF2 plays a major role in controlling mRNA transcription throughout the Drosophila genome to regulate genes functioning in cell growth, arguing that the term non-prototypic or specialized might not be entirely appropriate for this factor.
TAF3 anchors TFIID to nucleosomes
Trimethylation of lysine 4 on histone H3 (H3K4me3) at promoters is considered a mark of actively transcribed genes57,58. Timmers and colleagues discovered that the human TFIID complex binds H3K4me359, an interaction that is mediated by the TAF3 subunit, which contains a PHD finger that selectively binds H3K4me3 even when present in nucleosomes. Functional assays showed that the PHD finger of TAF3 can mediate transcriptional activation in a histone methyltransferase dependent fashion59. Other modifications on histone H3 can affect the association of TFIID with H3K4me3; asymmetric dimethylation of R2 inhibits binding of TFIID to H3 with the K4me3 mark, whereas acetylation of K9 and K14 augments TFIID binding to H3 with the K4me3 mark. The latter case is interesting, since H3K9Ac and H3K14Ac are associated with active promoters, and TAF1 has a double bromodomain known to bind acetyl-lysine, with a preference for diacetylated histone H460,61. These observations provide strong support for TFIID binding histone modifications in promoter regions, and highlight the possibility that TFIID is in part recruited to promoters containing specific histone modifications through the combined actions of TAF1 and TAF3. Perhaps the core promoter should be considered as not just DNA, but instead an ensemble of DNA elements and associated histones with specific modifications; it is this chromatin nucleoprotein complex containing the core promoter DNA that must be recognized by TFIID.
TBP bookmarks active genes during mitosis
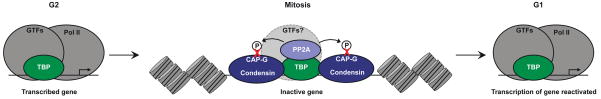
During mitosis transcription ceases, but the chromatin at the promoters of genes that were active prior to mitosis remains uncompacted or “bookmarked” so that when cells enter G1 these same genes again become transcriptionally active62. TBP, perhaps as part of TFIID, is involved in the process of bookmarking. Early studies showed that at least a portion of the TBP and TAFs remain bound to chromatin during mitosis, whereas other factors such as Pol II do not63–66. Recently, Sarge and colleagues further investigated the role of TBP in bookmarking (Figure 4)67. They found that in extracts from mitotic cells TBP binds both protein phosphatase 2A (PP2A) and the CAP-G subunit of condensin, the latter of which is a protein complex involved in compacting chromatin during mitosis. In these complexes, PP2A dephosphorylates CAP-G, which inhibits the activity of condensin. ChIP assays on mitotic cells showed that TBP and PP2A co-localize at the promoter of the histone H4 genes, which was active prior to mitosis. Decreasing TBP levels using an siRNA caused a substantial decrease in PP2A occupancy at the histone H4 promoter in mitotic cells, suggesting that TBP was required to maintain PP2A occupancy at this promoter during mitosis67. These observations lead to a model in which TBP recruits PP2A to the promoters of genes that are active prior to mitosis, where it phosphorylates CAP-G, thereby inhibiting the activity of condensin during mitosis. This prevents chromatin from condensing at these promoters, which bookmarks the promoters to become transcriptionally active when cells exit mitosis and enter G1.
Figure 4. TBP bookmarks genes during mitosis.
In mitotic nuclei, TBP (TATA binding protein) binds protein phosphatase 2A (PP2A) and the CAP-G subunit of condensin at the core promoters of genes that were active prior to mitosis67. PP2A dephosphorylates CAP-G, which inactivates condensin and keeps the core promoters from being tightly compacted in mitotic chromatin. This allows transcription of the bookmarked genes to be reactivated when cells exit mitosis and enter G1. During mitosis Pol II leaves chromatin and genes are transcriptionally inactive. Whether any general transcription factors (GTFs) other than TBP remain at bookmarked genes is unknown.
These observations raise many interesting questions. Is TBP in this context working as part of TFIID or in a different partnership with some other, as yet, undefined core promoter recognition complex? How many promoters require TBP for bookmarking and do they all contain TATA-boxes? Does TBP, which is also involved in RNA polymerase I and RNA polymerase III transcription, function in bookmarking promoters of genes that are transcribed by these polymerases? Do promoters of inactive genes that naturally harbor promoter-proximal paused polymerases also utilize TBP for bookmarking? Perhaps understanding the nature of the active preinitiation complex at the histone loci will provide insight into the mechanism of bookmarking.
Conclusions
Two of the unanticipated themes that have emerged from studies of non-prototypic core promoter recognition factors are: the roles of cell-type specific TAFs and TRFs in potentiating developmental gene regulation and cellular differentiation; and the apparent dispensability of canonical core promoter recognition components in some terminally differentiated mature cells that continue to actively synthesize cell and tissue specific mRNAs as was seen in the C2C12 model of skeletal muscle47,48. In the next few years, it will be interesting to see how many different mature cell types utilize these mechanisms to determine cell identity. It is possible that novel core promoter recognition factors might function to both guide cell determination and direct programs of gene expression in terminally differentiated adult cell-types. Indeed, recent studies suggest that loss of prototypic TFIID subunits and potential replacement by non-prototypic TAFs may be a prevalent mechanism that can be observed in both liver and fat cell differentiation in vitro and in vivo (J. D’Alessio, H. Zhou and R. Tjian, unpublished results). Future studies employing a combination of loss of function (shRNA depletion) or gain of function (ectopic expression) for non-prototypic core promoter recognition factors will shed light on this issue. For example, to gain more insight into the differential roles of variant core promoter machinery in different cell types, a combination of shRNA knockdown, mouse knockout, and ectopic expression of key factors (e.g. TRFs and TAFs) would be used. In some cases, one might even imagine that ectopic over expression of one set of TAFs/core factors may drive trans-differentiation of one precursor cell type into a different fate. Which cell systems and in vivo models will best serve to establish the physiological functions of cell specific core components is difficult to predict, but almost certainly a more fleshed out picture will require both purified cell types, in vitro biochemical systems, and animal studies. Clearly a significant challenge will be the availability of homogeneous and scalable cell types. Here, we expect that rapidly evolving single cell biochemistry technology and high-resolution imaging might provide a useful experimental avenue.
One can imagine that not all cell types will necessarily employ both of these mechanisms in the dramatic manner that has been observed for differentiated myotubes, which involves both the loss of TFIID and the retention and utilization of one select TAF (TAF3), possibly in conjunction with the non-prototypic TRF3. Indeed, we would expect that with hundreds, if not thousands of cell types in higher animals, there might very well be a variety of alternative mechanisms that take advantage of unique combinations of the TRFs and TAFs to work in concert with specific activators and repressors. When we drill down into multiple cell types to access their core transcription apparatus, we may see a complex palate of preinitiation complexes at work; in some cases the prototypic TFIID subunits may have become largely dispensable whereas in other cases select cell-type specifically expressed TAFs or TRFs may have assumed a more commanding role, while still functioning in collaboration with prototypical TFIID components and other cofactors such as Mediator. For example, in ovarian follicles, TAF4 is replaced by TAF4b in the mostly intact TFIID complex40. We might also imagine that in some mature cell types that may need to be rapidly reactivated to replicate (i.e. immune activation of B cells and T cells) TFIID levels may stay largely unchanged but one or more TAF subunit or TRF could become functionally more important.
It will be interesting to see whether some terminally differentiated cells that no longer replicate continue to use the prototypical TFIID as part of the preinitiation complex. In such cases, might these differentiated cells then modify the canonical core machinery and highjack their usual promoter recognition functions by substituting or adding new cell specific subunits, as has been documented in the case of TAF4b in the granulose cells of the mammalian ovary? In other situations where mature cells have exited the cell cycle, if one or two TAF/TRF factors are retained or even upregulated, while the bulk of the TFIID complex becomes largely eliminated, might different cell types enlist distinct sets of orphan TAFs and TRFs to meet their specific transcriptional needs? For example, instead of TAF3, which was used in myotubes, could some other orphan TAF become the key player in liver, fat, neurons, etc?
We might also imagine that the coordination between transcription factors of all types, including those we have described here, must dovetail with chromatin remodeling and modifying factors to initiate cell specific programs of transcription and maintain the specified differentiated state throughout the life of the mature cell. In this respect, we are particularly curious about the presence and activities of core promoter recognition factors during the formation of iPS cells. Here, terminally differentiated cells that in some case have likely jettisoned subunits of the prototypic TFIID complex must subsequently re-activate the myriad sets of genes encoding core promoter factors as well as DNA replication genes in order to become self renewing pluripotent iPS cells. Specifically, it will be interesting to learn the fate of TFIID, TAFs, and TRFs as distinct differentiated cells are forced to become iPS cells.
Hopefully, over the next several years, some of the questions raised above will be addressed, as core promoter recognition in specific cell types is elucidated using a more powerful repertoire of modern molecular and cell imaging tools, such as super high resolution microscopy for single cell analysis and in vitro biochemistry and cell-based assays in micro-fluidic chambers. If we are to understand what defines a cell type and how the many different functional cell types are derived, we must gain a more complete picture of the molecular players and transcriptional mechanisms controlling cell fate and identity. We anticipate that the core promoter recognition factors and functions discussed here as non-prototypic will themselves be considered prototypic as additional studies are performed in an expanding set of cells and differentiation pathways.
Presently, the mechanisms by which non-prototypic core machinery might direct cell specific programs of transcription have not been elucidated in any detail. A potential model might be reminiscent of the bacterial sigma-hypothesis, in which new core promoter recognition subunits of the holo-RNA polymerase can recognize and bind to a specific subset of promoters designated for sporulation or specific phage gene expression. It would not be difficult to imagine that novel TAF/TRF complexes can recognize and bind a distinct set of promoters containing unique, yet unidentified elements, and that various cell-type specifically expressed activators or repressors have evolved to target co-activator domains of these alternate core complexes as a means of expanding and diversifying the repertoire of combinatorial transactions. It is also possible that non-prototypic TAFs have the capacity to interact with specifically marked chromatin components and thereby regulate transcription activity by as yet unknown mechanisms.
Acknowledgments
We apologize to those whose relevant research we were unable to discuss because space limitations. J.G. was funded by Grant R01 GM55235 from the National Institute of General Medical Sciences and R.T. was partly funded by R37 CA25417 from the National Cancer Institute.
Glossary
RNA POLYMERASE II (POL II)
The enzyme that synthesizes mRNA in eukaryotic cells. Pol II is composed of 12 protein subunits (RPB1-RPB12). The binding of Pol II to promoters and the initiation of transcription requires a host of general transcription factors (TFII-A, B, D, E, F, H)
CORE PROMOTER
The region of a gene to which Pol II and the general transcription factors bind to initiate transcription. Core promoters span from approximately 40 base pairs upstream to 40 base pairs downstream of the transcription start site and are composed of DNA elements to which subunits of TFIID (or TFIIB) bind
PREINITIATION COMPLEX
The assembly of general transcription factors and Pol II on core promoter DNA. This complex, which can be assembled in the absence of nucleotide triphosphates in vitro, is competent to initiate transcription in the presence of nucleotides
CORE PROMOTER RECOGNITION FACTOR
A protein or multi-subunit complex that binds with sequence specificity to core promoter elements. The prototypical core promoter recognition factor for mRNA genes in eukaryotes is TFIID, subunits of which recognize multiple core promoter elements
TRANSCRIPTION FACTOR IID (TFIID)
A transcription factor for Pol II that binds core promoters. The TFIID complex is composed of the TATA binding protein (TBP) and 13-14 associated factors (TAFs)
TATA BINDING PROTEIN (TBP)
The central subunit of TFIID. TBP binds TATA boxes found in the core promoters of some eukaryotic mRNA genes
TBP ASSOCIATED FACTOR (TAF)
All subunits of the TFIID complex other than TBP are TAFs. There are 13-14 TAFs in the prototypical TFIID complex. There are also a number of proteins with sequence similarity to the prototypical TAFs, which are referred to as non-prototypic TAFs
TBP RELATED FACTOR (TRF)
A protein that is highly related in sequence to TBP. Two TRF proteins are discussed in this review – TRF2 (also known as TLF, TLP, TRP, or TBPL1) and TRF3 (also known as TBP2)
MITOTIC BOOKMARKING
The process by which genes that are active prior to mitosis are marked such that transcription begins again at these genes when cells exit mitosis and enter the G1 phase of the cell cycle
Contributor Information
James A. Goodrich, Email: james.goodrich@colorado.edu.
Robert Tjian, Email: jmlim@berkeley.edu.
References
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 3.Conaway RC, Conaway JW. Transcription initiated by RNA polymerase II and purified transcription factors from liver: transcription factors alpha, beta gamma, and delta promote formation of intermediates in assembly of the functional preinitiation complex. J Biol Chem. 1990;265:7559–7563. [PubMed] [Google Scholar]
- 4.Biggin MD, Tjian R. Transcription Factors That Activate the Ultabithorax Promoter in Develpmentally Staged Extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 5.Wasylyk B, Kedinger C, Corden J, Brison O, Chambon P. Specific in vitro initiation of transcription on conalbumin and ovalbumin genes and comparison with adenovirus-2 early and late genes. Nature. 1980;285:367–373. doi: 10.1038/285367a0. [DOI] [PubMed] [Google Scholar]
- 6.Horikoshi M, Carey MF, Kakidani H, Roeder RG. Mechanism of action of a yeast activator: direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell. 1988;54:665–669. doi: 10.1016/s0092-8674(88)80011-4. [DOI] [PubMed] [Google Scholar]
- 7.Lin YS, Carey MF, Ptashne M, Green MR. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 8.Carey M, Lin YS, Green MR, Ptashne M. A mechanism of synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Gralla JD, Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev. 1992;6:1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- 10.Goodrich JA, Hoey T, Thut CJ, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 11.Juven-Gershon T, Hsu JY, Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Biochem Soc Trans. 2006;34:1047–1050. doi: 10.1042/BST0341047. [DOI] [PubMed] [Google Scholar]
- 12.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2009;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu WL, et al. Structures of three distinct activator-TFIID complexes. Genes Dev. 2009;23:1510–1521. doi: 10.1101/gad.1790709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabenstein MD, Zhou S, Lis JT, Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci USA. 1999;96:4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teichmann M, et al. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci USA. 1999;96:13720–13725. doi: 10.1073/pnas.96.24.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore PA, et al. A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol Cell Biol. 1999;19:7610–7620. doi: 10.1128/mcb.19.11.7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maldonado E. Transcriptional functions of a new mammalian TATA-binding protein-related factor. J Biol Chem. 1999;274:12963–12966. doi: 10.1074/jbc.274.19.12963. [DOI] [PubMed] [Google Scholar]
- 18.Ohbayashi T, Makino Y, Tamura TA. Identification of a mouse TBP-like protein (TLP) distantly related to the drosophila TBP-related factor. Nucleic Acids Res. 1999;27:750–755. doi: 10.1093/nar/27.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science. 2001;292:1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]
- 20.Martianov I, et al. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell. 2001;7:509–515. doi: 10.1016/s1097-2765(01)00198-8. [DOI] [PubMed] [Google Scholar]
- 21.Martianov I, et al. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development. 2002;129:945–955. doi: 10.1242/dev.129.4.945. [DOI] [PubMed] [Google Scholar]
- 22.Kaltenbach L, Horner MA, Rothman JH, Mango SE. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell. 2000;6:705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 23.Veenstra GJ, Weeks DL, Wolffe AP. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science. 2000;290:2312–2315. doi: 10.1126/science.290.5500.2312. TRF2 was found to be highly expressed in Xenopus oocytes and embryos. Knock down of TRF2 showed that it is required for embryos to develop past the mid-blastula stage. This paper provided early evidence that non-prototypic core promoter recognition factors function in embryogenesis. [DOI] [PubMed] [Google Scholar]
- 24.Jacobi UG, et al. TBP paralogs accommodate metazoan- and vertebrate-specific developmental gene regulation. EMBO J. 2007;26:3900–3909. doi: 10.1038/sj.emboj.7601822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopytova DV, et al. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol Cell Biol. 2006;26:7492–7505. doi: 10.1128/MCB.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bashirullah A, Lam G, Yin VP, Thummel C. S dTrf2 is required for transcriptional and developmental responses to ecdysone during Drosophila metamorphosis. Dev Dyn. 2007;236:3173–3179. doi: 10.1002/dvdy.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persengiev SP, et al. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci USA. 2003;100:14887–14891. doi: 10.1073/pnas.2036440100. TRF3 was identified in the human, mouse, and frog genomes and the TRF3 protein was found expressed in many human and mouse tissues and cell lines. The identification of TRF3 set the stage for many other studies of this protein and its function in development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartfai R, et al. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 29.Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci USA. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao L, Kim M, DeJong J. Developmental and cell type-specific regulation of core promoter transcription factors in germ cells of frogs and mice. Gene Expr Patterns. 2006;6:409–419. doi: 10.1016/j.modgep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Gazdag E, Rajkovic A, Torres-Padilla ME, Tora L. Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggests different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction. 2007;134:51–62. doi: 10.1530/REP-06-0337. [DOI] [PubMed] [Google Scholar]
- 32.Gazdag E, et al. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes Dev. 2009;23:2210–2223. doi: 10.1101/gad.535209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. The protein encoded by the Drosophila Cannonball gene was found to be a homolog of TAF5. Cannonball is expressed in Drosophila testes where it controls expression of a series of genes in spermatocytes. After these studies were published additional testes specific TAFs were discovered that also function in spermatogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiller M, et al. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- 36.Dikstein R, Zhou S, Tjian R. Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. This paper documented the discovery of the first cell-type specific TAF, TAFII105 (now known as TAF4b), which is similar in sequence to TAF4. TAF4b was found associated with TFIID in a B cell line, but not in other cell lines. This paper raised awareness of the possibility of cell-type specific general transcription factors; subsequently, many other cell-type specific TAFs were discovered. [DOI] [PubMed] [Google Scholar]
- 37.Freiman RN, et al. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. Knockout of TAF4b in mice showed that it is required for development of the ovary. TAF4b is expressed in granulosa cells and is required for expression of ovarian-specific genes during folliculogenesis. [DOI] [PubMed] [Google Scholar]
- 38.Voronina E, et al. Ovarian granulosa cell survival and proliferation requires the gonad-selective TFIID subunit TAF4b. Dev Biol. 2007;303:715–726. doi: 10.1016/j.ydbio.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovasco LA, et al. Accelerated ovarian aging in the absence of the transcription regulator TAF4B in mice. Biol Reprod. 2010;82:23–34. doi: 10.1095/biolreprod.109.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geles KG, et al. Cell-type-selective induction of c-jun by TAF4b directs ovarian-specific transcription networks. Proc Natl Acad Sci USA. 2006;103:2594–2599. doi: 10.1073/pnas.0510764103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu WL, et al. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol Cell. 2008;29:81–91. doi: 10.1016/j.molcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falender AE, et al. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pointud JC, et al. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci. 2003;116:1847–1858. doi: 10.1242/jcs.00391. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Y, et al. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol. 2007;27:2582–2589. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. Knockout of TRF3 in zebrafish showed that it was required for hematopoiesis. TRF3 was found to bind the promoter of the mespa gene and to facilitate its expression. In the absence of mespa, hematopoiesis does not occur. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hart DO, Santra MK, Raha T, Green MR. Selective interaction between Trf3 and Taf3 required for early development and hematopoiesis. Dev Dyn. 2009;238:2540–2549. doi: 10.1002/dvdy.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. An unprecedented exchange of core promoter recognition machinery was found to occur when myoblasts differentiate into myotubes. Specifically, multiple subunits of TFIID were found to decrease, while levels of TAF3 remained constant. TAF3 binds TRF3 in myotubes and the two factors associate with the core promoter of the myogenin gene where they direct its transcription, thereby facilitating myogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deato MD, et al. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akhtar W, Veenstra GJ. TBP2 is a substitute for TBP in Xenopus oocyte transcription. BMC Biol. 2009;7:45. doi: 10.1186/1741-7007-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guermah M, Ge K, Chiang CM, Roeder RG. The TBN protein, which is essential for early embryonic mouse development, is an inducible TAFII implicated in adipogenesis. Mol Cell. 2003;12:991–1001. doi: 10.1016/s1097-2765(03)00396-4. [DOI] [PubMed] [Google Scholar]
- 51.Mohan WSJ, Scheer E, Wendling O, Metzger D, Tora L. TAF10 (TAF(II)30) is necessary for TFIID stability and early embryogenesis in mice. Mol Cell Biol. 2003;23:4307–4318. doi: 10.1128/MCB.23.12.4307-4318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tatarakis A, et al. Dominant and redundant functions of TFIID involved in the regulation of hepatic genes. Mol Cell. 2008;31:531–543. doi: 10.1016/j.molcel.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 53.Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- 54.Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007;21:2936–2949. doi: 10.1101/gad.1608807. TRF2 was found to direct the transcription of histone H1 genes, but not adjacent core histone genes, in the Drosophila histone gene cluster. TRF2 binds the promoter of the TATA-less histone H1 gene, whereas TBP binds the promoters of the core histone genes. Genome wide studies revealed that TRF2 widely binds TATA-less genes and drives their transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holmgren P, Johansson T, Lambertsson A, Rasmuson B. Content of histone H1 and histone phosphorylation in relation to the higher order structures of chromatin in Drosophila. Chromosoma. 1985;93:123–131. doi: 10.1007/BF00293159. [DOI] [PubMed] [Google Scholar]
- 56.Ruddell A, Jacobs-Lorena M. Biphasic pattern of histone gene expression during Drosophila oogenesis. Proc Natl Acad Sci USA. 1985;82:3316–3319. doi: 10.1073/pnas.82.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 58.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 59.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. TAF3 was found to bind a specific post-translationally modified form of histone H3 (H3K4me3) and to recruit TFIID to chromatin containing this modification. Importantly, H3K4me3 is a known mark for transcriptionally active genes. These studies lead us to reconsider the definition of a core promoter to include not just DNA elements, but also the specific post-translational modifications on histones associated with core promoter regions. [DOI] [PubMed] [Google Scholar]
- 60.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 61.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 62.Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 63.Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 64.Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- 65.Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell. 2002;13:276–284. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kieffer-Kwon P, Martianov I, Davidson I. Cell-specific nucleolar localization of TBP-related factor 2. Mol Biol Cell. 2004;15:4356–4368. doi: 10.1091/mbc.E04-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. TBP was found to function in mitotic bookmarking by binding PP2A and the CAP-G subunit of condensin at core promoters of genes active prior to mitosis. This assembly causes dephosphorylation of CAP-G, inactivation of condensin, and blocks the compaction of chromatin containing these core promoters. Once cells exit mitosis, transcription of the bookmarked genes is reactivated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimmins S, Kotaja N, Davidson I, Sassone-Corsi P. Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction. 2004;128:5–12. doi: 10.1530/rep.1.00170. [DOI] [PubMed] [Google Scholar]