Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases (original) (raw)
. Author manuscript; available in PMC: 2013 Nov 20.
Published in final edited form as: Inflamm Bowel Dis. 2010 Sep 13;17(1):10.1002/ibd.21339. doi: 10.1002/ibd.21339
Abstract
Background
Abnormal host-microbe interactions are implicated in the pathogenesis of inflammatory bowel diseases. Previous 16S rRNA sequence analysis of intestinal tissues demonstrated that a subset of Crohn’s disease (CD) and ulcerative colitis (UC) samples exhibited altered intestinal associated microbial compositions characterized by depletion of Bacteroidetes and Firmicutes (particularly Clostridium taxa). We hypothesize that NOD2 and ATG16L1 risk alleles may be associated with these alterations.
Methods
To test this hypothesis, we genotyped 178 specimens collected from 35 CD, 35 UC and 54 control patients for the three major NOD2 risk alleles (Leu 1007fs, R702W and G908R) and the _ATG16L1_T300A risk allele, that had undergone previous 16S rRNA sequence analysis. Our statistical models incorporated the following independent variables:1.) disease phenotype (CD, UC, Non-IBD Control); 2.) NOD2 composite genotype (NOD2R = at least one risk allele, NOD2NR = no risk alleles); 3.) ATG16L1T300A genotype (ATG16L1R/R, ATG16L1R/NR, ATG16L1NR/NR); 4.) patient age at time of surgery and all first order interactions. The dependent variable(s) were the relative frequencies of bacterial taxa classified by applying the RDP 2.1 classifier to previously reported 16S rRNA sequence data.
Results
Disease phenotype, NOD2 composite genotype and ATG16L1 genotype were significantly associated with shifts in microbial compositions by nonparametric MANCOVA. Shifts in the relative frequencies of Faecalibacterium and Escherichia taxa were significantly associated with disease phenotype by nonparametric ANCOVA.
Conclusions
These results support the concept that disease phenotype and genotype are associated with compositional changes in intestinal associated microbiota.
Keywords: NOD2, ATG16L1, inflammatory bowel diseases, microbiota
INTRODUCTION
The inflammatory bowel diseases (IBD) represent a heterogeneous spectrum of chronic inflammatory disorders of the digestive tract [1, 2]. Crohn’s disease (CD) and ulcerative colitis (UC) represent the two major IBD phenotypes. CD can affect any segment of the intestinal tract, often in a discontinuous fashion, but most commonly affects the distal ileum and colon. UC is confined within the colon, where disease extends proximally from the rectum in a continuous fashion.
Abnormal interactions between host and microbes (either pathogen or commensal) are implicated in the pathogenesis of IBD [3–8]. Studies of experimental animal models reveal that intestinal inflammation is not observed in germ-free mice. Antibiotics or diversion of fecal flow can reduce inflammation in experimental animal models and in humans in clinical settings. Moreover, both CD and UC are associated with significant shifts in the composition of the enteric microbiota (i.e., dysbiosis), most notably, depletion of the phyla Bacteroidetes and Firmicutes, compared to control patients [3–8]. For instance, we observed that the abundances of Bacteroidetes and Firmicutes were reduced 10–100 fold in surgically resected tissues collected from a subset of IBD patients, compared with control (non-IBD) patients [8]. The loss of Firmicutes was due primarily to reduction in the abundances of species that belong to the bacterial order clostridiales, particularly members of Clostridium clusters XIVa and IV [8,9].
Greater than forty inflammatory bowel disease susceptibility loci have been identified in the past 10 years through linkage analysis, association mapping, and candidate gene association studies [1, 10–18]. Risk alleles of two genes, NOD2 and ATG16L1, have been linked to ileal CD, a subphenotype of IBD, and to abnormalities in Paneth cell function [14,15,18]. Paneth cells are specialized small intestinal lining cells, with the highest concentration in the ileum, that secrete antimicrobial peptides and proteins, and thus potentially play an important role in host containment of lumenal bacteria. Disruption of normal Paneth cell function could therefore impact the composition and abundance of the intestinal microbiota, as has been observed in IBD. In order to test the hypothesis that human genetic factors are associated with shifts in microbial populations previously observed in a subset of inflammatory bowel disease intestinal samples [8], we genotyped these samples for the three major NOD2 (Leu1007fs, R702W, G908R) alleles and the _ATG16L1_T300A allele.
MATERIALS AND METHODS
Patients and acquisition of samples
This study was approved by the Institutional Review Boards of Washington University-St. Louis and Stony Brook University. The 178 de-identified intestinal tissue DNA samples were prepared as previously described from surgically resected tissues collected from 35 CD (both ileal and non-ileal CD), 35 UC and 54 control patients at the Mount Sinai School of Medicine under a protocol that was approved by the Institutional Review Boards of Mount Sinai Hospital and the University of Colorado [8]. These samples were linked to de-identified information on patient age at the time of surgery and disease phenotype. The diagnosis of CD or UC was established on the basis of gross and microscopic pathological features [8].
Genotyping of NOD2 and ATG16L1 SNPs
Genotyping of the three predominant NOD2 single nucleotide polymorphisms (SNPs),Leu1007fsInsC (rs2066847, SNP13), R702W (rs2066844, SNP8) and G908R (rs2066845, SNP12), and the ATG16L1T300A (rs2241880) SNP was conducted using the Sequenom MassArray System (Sequenom Inc., San Diego, CA) and/or the Taqman MGB (Applied Biosystems, Foster City, CA) genotyping platform according to the manufacturer’s recommendations [19]. For statistical analyses, risk alleles were combined into pooled genotypes. Thus, the NOD2 genotype categories were: 1.) no NOD2 risk alleles (NR/NR); 2.) one NOD2 risk allele (R/NR); and 3.) two NOD2 risk alleles (R/R). Similarly, the ATG16L1 genotype categories were 1.) no ATG16L1 risk allele (NR/NR); 2.) one ATG16L1 risk allele (R/NR);and 3.) two ATG16L1 risk allele (R/R).
Statistical analysis
The bacterial rRNA sequences (depth of ∼80/sample) of the intestinal samples have been previously reported [8]. The previous phylogenetic classification of rRNA sequences was confirmed using the Naïve Bayesian Classifier of the Ribosome Database Project (Version 2.1, http://sourceforge.net/projects/rdp-classifier/) [20,21]. The taxa were binned at the phyla level: 1. Actinomycetes; 2. Bacteroidetes; 3. Firmicutes; 4. Proteobacteria; 5. Other phyla. The Firmicutes clade was further subdivided into the following five subcategories, based on concordance between the RDP classifier and the Greengenes 16S rRNA phylogenetic schema [8,20–22]: 3A. Firmicutes/Clostridium Group XIVa, 3B. Firmicutes/Clostridium Group IV, 3C. Firmicutes/Clostridium/ Other, 3D. Firmicutes/Bacillus, and 3E. Firmicutes/Other classes. The relative frequencies of each of the eight categories were subjected to logit transformation: log2{(x+0.01)/[100-(x+0.01)]}, where x is the relative frequency (%). The effect of clinical phenotype (CD, UC, Control), NOD2 genotype (NR/NR, R/NR, R/R),ATG16L1 genotype (NR/NR, R/NR, R/R), and patient age on these eight categories was analyzed by nonparametric multivariate analysis of covariance (MANCOVA) [23,24] using the R software package (Version 2.8.1). The purpose of the logit transformation was to expand the original relative frequency values (0–100%) to the real space, in which the term 0.01 was added to avoid infinite value during the transformation. The effect of these independent variables on lower taxonomic ranks was analyzed by nonparametric analysis of covariance (ANCOVA) [23,24].
UniFrac tests [25] were performed using the web-service provided at http://bmf.colorado.edu/unifrac. The phylogenetic tree tested in this analysis was constructed by neighbour joining of sequences representative of 95% operational taxonomic units (OTUs; assembled through the application sortx [26], followed by parsimony insertion of remaining sequences, by use of the ARB software package [26]. UniFrac significance and environmental clustering was ascertained by 100 jackknife re-samplings using normalized, weighted UniFrac. Dendrograms were constructed from UniFrac scores through Unweighted Pair Group Method with Arithmetic (UPGMA) mean clustering.
RESULTS
The distribution of NOD2 and ATG16L1 genotypes in 178 tissue samples are shown in Table 1. Because some combinations of NOD2, ATG16L1, and disease phenotype were not sampled in this study, we collapsed the NOD2 genotypes into two categories: NOD2R (harboring at least one of the three major NOD2 risk alleles) and NOD2NR (harboring none of the three major risk alleles). We retained the three ATG16L1T300A genotype categories: R/R, R/NR, and NR/NR. In our statistical model we incorporated the three disease phenotypes: 1.) CD and 2.) UC and 3.) Control non-IBD samples (collected predominantly from cancer patients; [8]). As previously reported [8], the CD and UC patients were significantly younger at the time of surgery than the control patients.
Table 1.
Sample characteristics with respect to disease phenotype, NOD2 genotype, ATG16L1 genotype and age.
| CD_n = 61_ | UC_n = 58_ | Control_n = 59_ | |
|---|---|---|---|
| NOD2R/R ATG16L1R/R | 0.0% | 3.4% | 1.7% |
| NOD2R/R ATG16L1R/NR | 4.9% | 0.0% | 0.0% |
| NOD2R/R ATG16L1NR/NR | 3.3% | 0.0% | 0.0% |
| NOD2R/NR ATG16L1R/R | 24.6% | 0.0% | 6.8% |
| NOD2R/NR ATG16L1R/NR | 8.2% | 10.3% | 3.4% |
| NOD2R/NR ATG16L1NR/NR | 3.3% | 0.0% | 3.4% |
| NOD2NR/NR ATG16L1R/R | 16.4% | 20.7% | 20.3% |
| NOD2NR/NR ATG16L1R/NR | 23.0% | 34.5% | 28.8% |
| NOD2NR/NR ATG16L1NR/NR | 16.4% | 31.0% | 35.6% |
| Median age years (range) | 40y (18–68) | 40y (19–69) | 64y (25–89) |
The previously reported 16S sequencing data for these 178 specimens [8] were reclassified using the recently released Ribosomal Database Project (RDP) 2.1 classifier [20,21]. This software assigned >98% of the sequences to one of four phyla: Actinobacteria, Bacteroidoetes, Firmicutes and Proteobacteria. The Firmicutes clade was further divided to correspond to Clostridium Group XIVa and IV groups [8, 9,20,21]. As shown in Table 2, the relative frequencies of Clostridium Group XIVa and Clostridium Group IV taxa were significantly decreased in CD compared to Control samples, and the relative frequencies of Actinobacteria and Proteobacteria taxa were significantly increased. Nonparametric MANCOVA analysis of the effect of clinical phenotype, NOD2 composite genotype, ATG16L1 genotype, and age of surgery on intestinal (ileum and colon) microbial composition, as defined by the relative frequencies of 8 bacterial categories (excluding Other Phyla), revealed that clinical phenotype, NOD2 composite genotype, and ATG16L1 genotype were associated with shifts in microbial composition (Table 3).
Table 2.
The relative frequencies of eight bacterial categories.
| Taxon | CDn = 56 | UCn = 54 | Controln = 58 |
|---|---|---|---|
| Actinobacteria | 6.8 ± 13.3* | 5.7 ± 7.7 | 3.7 ± 8.1 |
| Bacteroidetes | 19.1 ± 21.4 | 21.8 ± 23.6 | 26.1 ± 23.1 |
| Firmicutes/Clostridium Group XIVa | 11.1 ± 15* | 17.9 ± 20.3 | 22.9 ± 21.6 |
| Firmicutes/Clostridium Group IV | 4.5 ± 9.8* | 6.3 ± 8.8 | 9.7 ± 11.9 |
| Firmicutes/Clostridium/ Other genera | 5.6 ± 17.4 | 5.0 ± 7.6 | 6.1 ± 10.5 |
| Firmicutes/Bacilli | 21.0 ± 22.2 | 18.9 ± 24.8 | 19.9 ± 30.0 |
| Firmicutes/Other classes | 0.1 ± 0.3 | 0.2 ± 0.5 | 0.7 ± 2.9 |
| Proteobacteria | 31 ± 27.8* | 23.1 ± 25.0 | 9.5 ± 19.0 |
| Other phyla | 0.9 ± 2.9 | 1.3 ± 4.9 | 1.5 ± 5.7 |
Table 3.
Non-parametric MANCOVA results.
| Source | DF | F value | Pr > F |
|---|---|---|---|
| Disease phenotype | 2,158 | 5.532 | 0.002 * |
| NOD2 (composite) genotype | 1,158 | 2.756 | 0.024 * |
| ATG16L1 genotype | 2,158 | 2.637 | 0.011 * |
| Age | 1,158 | 2.173 | 0.055 |
| NOD2 genotype : disease phenotype | 2,158 | 1.057 | 0.36 |
| NOD2 genotype : ATG16L1 genotype | 2,158 | 0.795 | 0.603 |
| NOD2 genotype : age | 1,158 | 6.579 | 0.001* |
| ATG16L1 genotype : disease phenotype | 4,158 | 1.693 | 0.052 |
| ATG16L1 genotype : age | 2,158 | 1.782 | 0.077 |
| Disease phenotype : age | 2,158 | 1.019 | 0.385 |
To determine whether particular lower-level taxonomic groups (i.e., sub-phylum) were associated with clinical phenotypes or risk alleles, nonparametric ANCOVA analyses were conducted on individual genera within Clostridial clusters XIVa (e.g., Lachnospiracea) and IV (e.g., Ruminococcaceae). Indeed, the relative abundances of several clostridial genera were associated with disease phenotype, NOD2 composite genotype, and/or ATG16L1 genotype (Table 4). Within the phylum Proteobacteria, only the genus Escherichia was significantly populated in all the specimens (See Table 4). Association between disease phenotype and shifts in the Faecalibacterium and the Escherichia remained significant after applying the Bonferroni correction for multiple comparisons (n=11).
Table 4.
Nonparametric ANCOVA results for individual genera.
| Taxon | Diseasephenotype | NOD2compositegenotype | ATG16L1genotype | Age ofsurgery |
|---|---|---|---|---|
| “Firmicutes”/”Clostridia”/”Clostridiales”/”Lachnospiraceae” | ||||
| Coprococcus | 0.011 | 0.127 | 0.043 | 0.251 |
| Dorea | 0.024 | 0.025 | 0.087 | 0.428 |
| Roseburia | 0.013 | 0.928 | 0.157 | 0.586 |
| Syntrophococcus | 0.103 | 0.809 | 0.034 | 0.913 |
| “Firmicutes”/”Clostridia” Clostridiales”/“Ruminococcaceae” | ||||
| Anaerotruncus | 0.387 | 0.899 | 0.015 | 0.471 |
| Butyricicoccus | 0.006 | 0.807 | 0.138 | 0.79 |
| Faecalibacterium | 0.001* | 0.523 | 0.018 | 0.363 |
| Oscilibacter | 0.028 | 0.069 | 0.176 | 0.096 |
| Ruminococcus | 0.042 | 0.946 | 0.187 | 0.253 |
| Subdoligranulum | 0.087 | 0.083 | 0.007 | 0.298 |
| “Proteobacteria”/Gamma proteobacteria/“Enterobacteriales”/Enterobacteriaceae | ||||
| Escherichia | 0.001* | 0.96 | 0.397 | 0.013 |
The aforementioned statistical modeling analyzed the relative abundances of sequence relatedness groups (operational taxonomic units) that were assembled on the basis of taxonomic sequence classification. Phylum and sub-phylum level sequence classifications using the Naïve Bayesian classifier were nearly identical to our previously reported groupings. Thus the reported statistical associations between clinical phenotypes, risk allele genotypes, and intestinal microbiotas are robust to particular schemes of OTU classification. Nevertheless, we sought to provide a non-OTU-based assessment of these results through comparison of phylogenetic community structure.
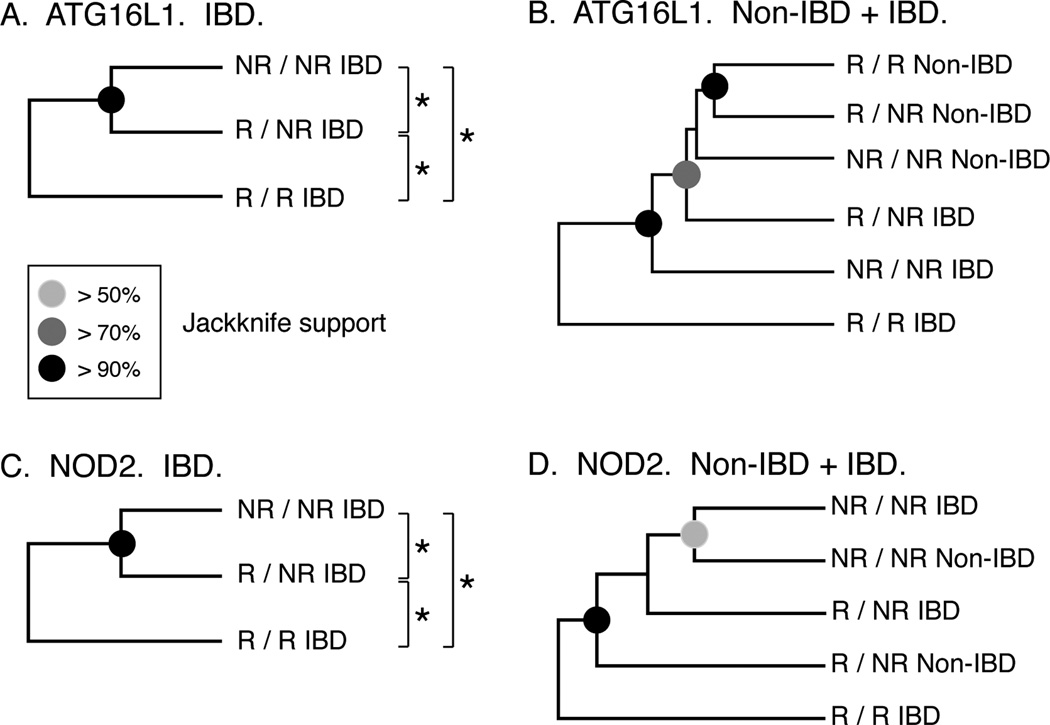
In general, analysis of microbial communities using the UniFrac metric [25] corroborated the findings that ATG16L1 and NOD2 risk alleles are associated with altered microbiotas. To mask potential confounding by disease phenotype, we first compared the microbiotas of IBD patients as a function of ATG16L1 and NOD2 risk alleles taken independently (Fig. 1A and 1C). In the case of each gene, the microbiotas of NR/NR, NR/R, and R/R subjects differed significantly from one another (P < 0.03 for each pairwise comparison). Distance-based clustering of microbiotas indicated that the microbiotas of NR/NR and NR/R individuals were more similar to one another than either one were to R/R subjects (Jackknife support ≥ 90%). Thus, an allele-dosage effect on the intestinal microbiotas was apparent in the analyses of both ATG16L1 and NOD2. R/R IBD subjects also were observed to be outliers when non-IBD controls were included in the analysis (Fig. 1B and 1D). Interestingly, for ATG16L1, non-IBD microbiotas were observed to cluster to the exclusion of IBD microbiotas, indicating that disease phenotype (IBD vs. Non-IBD) was a critical modifier of the microbiota. In contrast, NOD2 genotypes tended to cluster together, regardless of disease phenotype (none of the Non-IBD control subjects had a NOD2 R/R genotype). However, none of the pair-wise comparisons of genotype/phenotype categories was statistically significant when P-values were corrected for multiple comparisons.
DISCUSSION
The objective of this study was to determine whether human genetic factors underlie shifts in microbial populations that have been observed in a subset of CD and UC patients [8]. We focused on NOD2 and ATG16L1 risk alleles, which are associated with abnormal Paneth cell function. The NOD2 gene is constitutively expressed in human Paneth cells [14], which are specialized intestinal cells that secrete antimicrobial peptides and proteins into intestinal crypts. In one study [15], reduced α-defensin expression was associated with NOD2 risk alleles. Although the ATG16L1 gene is expressed in multiple cell lineages, the ATG16L1 risk allele has been associated with abnormalities in both mice and human Paneth cells [18]. We reason that alterations in Paneth cell function can perturb host containment of lumenal bacteria, particularly in the ileum where Paneth cells are most abundant [28]. This concept has been supported by the finding that genetic mouse models with altered secretion of enteric defensins from Paneth cells exhibit alterations in small intestinal microbial composition [29].
Analysis of the effect of NOD2 and ATG16L1 risk alleles on the intestinal microbiota is complicated by established associations between the NOD2 and ATG16L1 risk alleles with disease subphenotype (ileal CD but not Crohn’s colitis) and earlier age of surgery. For this reason, all of these variables were included in the multivariate analyses. We confirmed that decreases in the relative frequencies of Clostridial XIVa and IV taxa and increases in Proteobacteria taxa were observed in CD specimens compared to control specimens [3,7,8]. Our results support the hypothesis that both genetic factors and disease phenotype are associated with significant shifts in microbial populations.
Analysis of bacterial genera within the phyla Firmicutes and Proteobacteria confirmed previous results linking shifts in Faecalibacterium and Escherichia with disease phenotype [30,31]. Inspection of the data also suggests that shifts in the relative frequencies of several genera may be associated with NOD2 and/or ATG16L1 genotype, but these results did not reach significance after correction for multiple comparisons. However given the shallow level of DNA sequence coverage in this dataset, which resulted in low observed sequence abundances for most genera, it is likely that analyses of individual genera were underpowered. Studies of monozygotic twins that were either discordant or concordant for CD have shown that changes in microbial composition are associated with development of ileal CD subphenotype [31]. Thus the observed shifts in microbial populations could potentially reflect simply the presence of ileal CD.
The 178 specimens analyzed in this study were heterogeneous with respect to CD subphenotypes because ileal CD and Crohn’s colitis patients were not differentiated in this study. Specimens were collected from both the ileum and colon, and included samples of both grossly disease unaffected and affected regions of the intestine (gross pathology was not significantly associated with alterations in microbial populations in our initial study; [8]). Furthermore, because surgical histories were not recorded, the contribution of previous surgeries (e.g. previous ileocolic resections) to shifts in microbial composition cannot be evaluated in this study. Further analysis of the contribution of ATG16L1 risk allele and other CD risk alleles to shifts in mucosal associated microbiota as it relates to IBD pathogenesis, will require comprehensive analysis of additional patient samples with detailed annotation of disease phenotype (including CD subphenotypes), history of previous surgeries, genotype, anatomic location, pathology (restrict to disease unaffected regions), as well as more extensive analysis of other microbiota subsets.
Despite potential confounding variables, the results of this study strongly suggest that specific genetic loci implicated in human disease can impact the composition of the intestinal microbiota. Although imbalances in commensal microbiota have been associated with many human disease states (CD, obesity, antibiotic-associated diarrhea, prostatitis), the causal relationships between disease and dysbiosis remain unclear in most cases. However, our observation that shifts in microbial populations are associated with particular CD risk alleles indicates that dysbiosis is not simply a consequence of a chronic disease and its attendant, long-term treatment. Additional experimental and clinical studies are necessary to determine whether and how altered commensal populations contribute to disease progression and resolution.
FIGURE 1.
UniFrac analysis of CD risk alleles and enteric microbiota. The images show the results of UPGMA clustering of environments (A). ATG16L1 genotype in IBD patients (B). NOD2 genotype in IBD patients (C). ATG16L1 genotype in IBD and non-IBD control patients (D). NOD2 genotype in IBD and non-IBD control patients based on the UniFrac metric.24 Jackknife resampling (100 replicates) was performed to assess the statistical support for the topology of each tree. Nodes with high support are indicated by the fraction of trees in which the nodes were recovered (>50%: >50% to <70%; >70%: >70% to <90%; >90%: >90% to 100%).
ACKNOWLEDGEMENTS
The authors wish to thank Dr. Chuck Elson for suggesting this experiment.
This work was supported by NIH grants UH2DK083994 (Li), contract no. NO1-AI-30055 (Boedeker), a grant from the Crohn’s and Colitis Foundation of America (Li) and a grant from the Simons Foundation (Li). We acknowledge use of the Washington University Digestive Diseases Research Core Center Tissue Procurement Facility (P30 DK52574). This work was presented in part at the American Gastroenterology Association meeting held in Washington D.C., May 2009.
REFERENCES
- 1.Abraham C, Cho J. Mechanisms of disease: inflammatory bowel disease. New Engl. J Medicine. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satsangi J, Silverberg MS, Vermiere S, Colombel J-F. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–753. doi: 10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 4.Eckburg PB, Relman DA. The role of microbes in Crohn’s disease. Clin Infect. Dis. 2007;454:256–262. doi: 10.1086/510385. [DOI] [PubMed] [Google Scholar]
- 5.Elson CO, Cong Y, McCracken VJ, et al. Experimental models of inflammatory bowel disease reveal innate, adaptive and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 6.Peterson DA, Frank DN, Pace N, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokol H, Lay C, Seksik P, Tannock GW. Analysis of bacterial bowel communities of IBD patients: What has it revealed? Inflamm Bowel Dis. 2008;14:858–867. doi: 10.1002/ibd.20392. [DOI] [PubMed] [Google Scholar]
- 8.Frank DN, St.Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins MD, Lawson PA, Willems A, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 10.Goyette P, Labbé C, Trinh TT, et al. Molecular pathogenesis of inflammatory bowel disease: genotypes, phenotypes and personalized medicine. Ann Med. 2007;39:177–199. doi: 10.1080/07853890701197615. [DOI] [PubMed] [Google Scholar]
- 11.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 13.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 14.Ogura Y, Lala S, Xin W, et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut. 2003;52:1591–1597. doi: 10.1136/gut.52.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wehkamp J, Harder J, Weichenthal M, et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prescott NJ, Fisher SA, Franke A, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007 May;132(5):1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 18.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008 Nov 13;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abreu MT, Taylor KD, Lin YC, et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn’s disease. Gastroenterology. 2002;123:679–688. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole JR, Wang Q, Cardenas E, Fish J, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 24.McArdle BH, Anderson MJ. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- 25.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank DN. XplorSeq: a software environment for integrated management and phylogenetic analysis of metagenomic sequence data. BMC Bioinformatics. 2008;9:420. doi: 10.1186/1471-2105-9-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig W, Strunk O, Westram R, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzman NH, Underwood MA, Bevins CL. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19:70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Salzman NH, Hung K, Haribhai D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sokol H, Pigneur B, Watterlot L, Lakhdari O, et al. Faecalibacterium prausnitzii is an anti inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willing B, Halfvarson WB, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosal-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]