NUT Midline Carcinoma (original) (raw)
. Author manuscript; available in PMC: 2011 Nov 1.
Abstract
NUT midline carcinoma (NMC) is a rare, genetically defined, aggressive human cancer defined by rearrangements of the gene NUT. In the majority (approximately 75%) of NMCs most of the coding sequence of NUT on chromosome 15q14 is fused BRD4 or BRD3, creating chimeric genes that encode BRD-NUT fusion proteins. In the remaining cases, NUT is fused to an unknown partner gene; these tumors are termed NUT-variant. Diagnosis of NMC is made by demonstration of expression of the NUT-fusion protein using a monoclonal antibody to NUT for immunohistochemistry, and confirmation of the fusion (BRD-NUT or NUT-variant) by fluorescent in situ hybridization or RT-PCR. Because the reagents and expertise required to diagnose NMC are not available in most laboratories, and the incomplete awareness of this disease, NMC is frequently undiagnosed or misdiagnosed, and its actual prevalence is unknown. NMC does not arise from any specific tissue type or organ. It presents as a poorly differentiated carcinoma originating from midline locations such as the head, neck or mediastinum. Although rare, NMCs occur throughout life and advanced local disease is frequently accompanied by distant hematogenous metastases. There remains no effective treatment for NMC, there are no guidelines, and current approaches to treatment are based on discussions amongst a few oncologists who each have had a single experience treating this disease. BRD-NUT functions to block cellular differentiation and promote uncontrolled growth of carcinoma cells.
Keywords: BRD4, NUT, BRD4-NUT, t(15; 19), carcinoma
History
In the summer of 1999, a 12 year old girl, who was to become the first American patient with NUT midline carcinoma, developed a sore throat and difficulty swallowing, and eventually developed a muffled voice. There was no response to antibiotics, and a month later was seen by an otolaryngologist. Upon examination, an ulcerating mass was found to have replaced her epiglottis. The biopsy pathology resembled that of a nasopharyngeal carcinoma. She was treated accordingly, at Children’s Hospital, Boston, and enjoyed an initial shrinkage of her tumor, but the tumor came back and she died a horrible death, strangled by the tumor which closed off her airway. The clinicians would not forget this tumor, but the abrupt end to her life marked the discovery of a new type of cancer.
It was fortunate that the pathologists at Children’s Hospital had sent some of the patient’s initial biopsy to Dr. Paola Dal Cin of Brigham and Women’s Hospital for cytogenetic analysis. She found that this was no ordinary carcinoma, a class of solid tumors characterized by notoriously complex karyotypes. Apart from a trisomy 8, the patient’s tumor’s 47 xx karyotype harbored one distinct abnormality, a t(15;19)(q13;p13.1). The news of this karyotype was noticed by Dr. Jonathan Fletcher, who pays attention to rare, translocation-associated solid tumors. His review of the literature revealed the existence of three isolated case reports of pediatric thymic carcinomas with associated t(15;19)s, all aggressive and rapidly lethal, a clinical course which resembled that of the index patient [1]. From one of these cases, which originated in Japan, Dr. Kubonishi had created a cell line [2], Ty-82. With the purpose of cloning the presumed fusion oncogene resulting from the t(15;19) that was hypothetically responsible for creating this unusual and aggressive carcinoma, Dr. Fletcher acquired the cell line and handed over the gene mapping project to a very green yet eager post-doctoral fellow, this author.
Breakpoint mapping back then, 2000, had been made much easier with the newly available YAC and cosmid libraries that could be used to FISH-map rapidly. This was done for both the 15q13 and 19p13.1 loci in less than a year, and the loci were finely mapped by southern analysis. The chromosome 19 locus was located in the middle of a newly described gene, BRD4 [3], and the chromosome 15 locus appeared to be very close to a small gene, Nop10p. Attempts to demonstrate that the Nop10p gene was present in a fusion transcript with BRD4 failed, however, and around that time a few new ESTs appeared within the area on the UCSC genome browser. Having failed to RACE out a product, we attempted RT-PCR using primers to these novel ESTs, and quickly succeeded at amplifying an in-frame fusion transcript between BRD4 and these ESTs. The ESTs collectively formed a putative 3.5kb gene that we initially called “PRUC”, for proline-rich in undifferentiated carcinoma. Unimpressed with this term, and having observed that it appeared to be expressed only in testis, as demonstrated in multiple tissue northern blotting [4], we chose a more appropriate acronym, NUT, for Nuclear protein in testis. That name has since been changed to “chr15orf55” by the concensus of nomenclature rules, but we still use the name NUT. Hence, the name BRD4-NUT for this fusion oncogene remains unchanged.
In the course of FISH-mapping the chromosome 19 and 15 breakpoints, we had developed some robust probes that could be used to screen new and archival tumors in search of more cases of what we termed, “t(15;19) carcinomas”. Our criteria were broad. We looked at all poorly differentiated carcinomas in patients younger than forty years of age, ignoring, for the time, adult carcinomas which were predicted to harbor 100s of mutations and complex karyotypes, a pathogenetic mechanism unlikely to overlap that of the t(15;19) carcinoma. We found seven tumors with NUT rearrangements within 98 tumors screened [5]. What was interesting was that while four of these were BRD4-NUT tumors, three had rearrangement of NUT, but not BRD4, termed NUT-variants. This has led to the hypothesis that the common denominator was NUT, and not BRD4. Hence, after several iterations, the current name of this cancer is NUT midline carcinoma, or NMC, to reflect that it is a defined by chromosomal rearrangement of NUT.
Clinico-pathologic features
To date, we have collected enough cases of NMC, screened mostly by FISH, to be able to roughly describe the disease as follows. While likely rare, the actual frequency of NMC has not been determined. As above, 7% of 98 mostly carcinomas in patients less than 40 were NMCs [5]. In a more recent study that was not restricted to young adults and children, NMC made up 18% of poorly differentiated carcinomas of the upper aerodigestive tract, particularly the sinonasal region [6]. Importantly, the average age of patients with NMC in this study was 47, overturning the perception that NMCs are largely confined to children and young adults [5]. Parenthetically, many recent patients in this author’s experience have been adults over the age of 30.
The clinico-pathologic features of NMCs are summarized in table 1 [7]. The age at presentation ranges from 3 to 78 years. The young age (avg. age 25) is likely distorted by selection bias in the original study as described above [5]. NMC is a genetically defined disease and does not arise from a specific organ. Most cases occur in the mediastinum and upper aerodigestive tract, but exceptional tumors have arisen in bone [16], the bladder [5], the abdominal retroperitoneum, the pancreas, and the salivary glands [17, 18].
Table 1.
Clinical characteristics of reported NUT midline carcinomas (from [7]).
| Caseno. | Age(yr.) | sex | Location | Diagnosis | Survival(mo.) | Brd4-NUT | NUT-variant | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | M | Bladder | PD sq cell ca | 8.5 | √ | [5] | |
| 2 | 5 | M | Mediastinum | Mucoepidermoid ca | 3.5 | √ | [8] | |
| 3 | 10 | M | Iliac bone | Ewing’s sarcoma/PNET | AW/OD 180* | √ | [9] | |
| 4 | 11 | F | Thorax | Undiff. ca | 4.5 | √ | [10] | |
| 5 | 12 | F | Nasopharynx | PD sq cell ca | 3.25 | √ | [1, 11] | |
| 6 | 13 | F | Epiglottis | Undiff. ca | 9 | √ | [1] | |
| 7 | 15 | M | Mediastinum | Thymic PD sq cell ca | 6 | √ | [12] | |
| 8 | 15 | F | Orbit | PD sq cell ca | 7 | √ | [5] | |
| 9 | 16 | M | Lung | Sq cell ca | 37 | √ | [5, 13] | |
| 10 | 16 | F | Trachea | PD ca | 31.5 | √ | [5] | |
| 11 | 21 | F | Nasopharynx | Nasopharyngeal ca | 10.25 | √ | [5] | |
| 12 | 22 | F | Thymus | PD ca | 3.5 | √ | [14] | |
| 13 | 26 | M | Sinonasal | Undiff ca | 16.75 | √ | [5] | |
| 14 | 30 | F | Mediastinum | PD ca | 3.25 | √ | [15] | |
| 15 | 31 | M | Nasal Cavity | SNUC | √ | |||
| 16 | 34 | F | Thorax | NK | NK | √ | [9] | |
| 17 | 34 | M | Mediastinum | PD ca | 2 | √ | [13] | |
| 18 | 35 | F | Mediastinum | PD ca | 2 | √ | [5] | |
| 19 | 39 | F | Nasal cavity and sinus | PD sq cell ca | √ | |||
| 20 | 40 | F | Nasal cavity and sinus | PD sq cell ca | √ | |||
| 21 | 47 | M | Nasal cavity and sinus | SNUC | √ | |||
| 22 | 78 | F | Supraglottic larynx | Undiff ca | √ | |||
| Avg. | 25.14 | 9.53 |
With the exception of one case [16], all patients with NMCs have died (table 1). The average survival is less than one year (9.5 mo.) despite aggressive chemotherapy and radiation treatment. Some data [5] and unpublished observations suggests that patients with _NUT_-variant NMCs may live longer than those with BRD4-NUT rearrangements (cases 9–11, table 1); this has yet to be formally demonstrated in a larger series.
Molecular Cytogenetics
Perhaps the most unique feature of NMCs are their simple karyotypes. The tumors often harbor only a single abnormality, the t(15;19)(q14;p13.1), and in this way more resemble leukemia than carcinoma, again pointing to the likely critical biologic importance of the fusion oncogene. Variants which create the BRD4-NUT fusion have included more complex three-way translocations [12] and unpublished observations, and some tumors have had cryptic NUT breakpoints undetectable by conventional FISH [19].
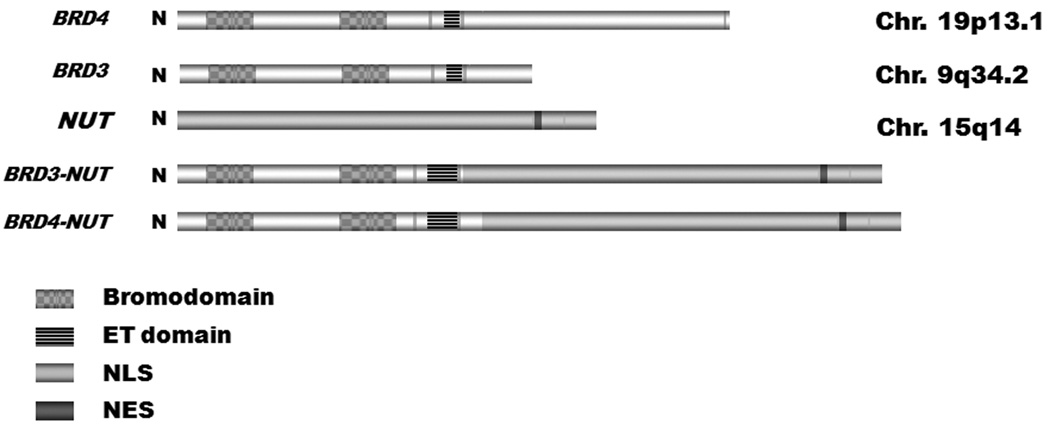
In approximately 2/3rds of cases, NUT (chromosome 15q14) is fused to BRD4, on chromosome 19p13.1, forming the BRD4-NUT fusion gene (fig 1) [4]. The remaining 1/3rd of cases are NUT-vairants where the partner gene is BRD3 or other uncharacterized genes [20]. The NUT promoter is active only in adult testis and ciliary ganglion [4, 20], and as a result, only one of the two fusion genes (e.g., BRD-NUT, but not NUT-BRD) are expressed. The breakpoints themselves characteristically occur within intron 10 of BRD4, or intron 9 of BRD3, and intron 2 of NUT, fusing the business end of BRD4, encoding both acetyl-histone binding bromodomains and extraterminal domain, with virtually the entire NUT gene.
FIG 1.
Cartoon of BRD4, BRD3, NUT, and BRD-NUT fusion proteins. ET, extraterminal domain. NLS, nuclear localization signal. NES, nuclear export signal.
Diagnosis
The histologic features of NMC are, unfortunately, not diagnostic. The morphology is that of a poorly differentiated carcinoma, with or without squamous differentiation. It does have a distinctly monomorphic, clonal appearance, as contrasted with the garden variety poorly differentiated carcinoma, which tends to be pleomorphic. NMC is a new disease, and is not broadly known to most pathologists nationally and internationally. It is therefore commonly undiagnosed, or misdiagnosed, most often as squamous cell carcinoma, but occasionally as other specific pathogenetic entities, such as Ewing sarcoma or Sinonasal undifferentiated carcinoma (SNUC). The result is that the actual frequency of NMC is unknown, and likely much more common than currently thought to be.
The diagnosis of NMC has historically been made by demonstration of NUT rearrangement by dual color, split-apart FISH using probes flanking NUT, or by demonstration of a BRD4-NUT fusion transcript by RT-PCR [15]. FISH is preferred because it will detect all NMCs, including all _NUT_-variants, whereas RT-PCR can currently only detect _BRD3_- or BRD4-NUT tumors. Because these assays are generally not available in most pathology or molecular diagnostic laboratories, we sought to develop a diagnostic monoclonal antibody to NUT, taking advantage of the fact that the native protein is not expressed outside of the testis. Staining a large panel of common carcinomas (n~1000), and including 30 FISH-positive NMCs, with the NUT antibody by immunohistochemistry, we found that it had a sensitivity of 87%, and a specificity of 100% [19]. We now use this antibody (Cell Signaling Technologies, Inc., Danvers, MA) routinely for diagnosis. The advent of this new immunohistochemical test for NMC makes diagnosis easy in any pathology laboratory, and due to the new user-friendliness of the diagnosis, it is expected that NMC will be added to the differential diagnosis of all non-smoking-related poorly differentiated carcinomas. Consequently, a much more accurate measure of the frequency of this disease is predicted to emerge in the near future.
Now, the challenge is not the diagnosis of NMC, but when to perform the immunohistochemical test for NUT expression. A useful guideline is to consider NMC in the differential diagnosis of any midline poorly differentiated carcinoma in a never-smoking patient, and in the absence of EBV or HPV within the tumor.
BRD4, NUT, and BRD4-NUT Function
BRD4 is known to bind acetylated histones, which are associated with actively transcribed DNA, including those of mitotic chromosomes [21, 22]. Through its constitutive association with active chromatin, as well as with the transcriptional elongation complex, pTEFb, BRD4 is thought to preserve cellular memory by marking regions for re-initiation of transcription following mitosis [23–26]. NUT encodes an unstructured polypeptide expressed only in post-meiotic spermatids, but whose function is otherwise unknown.
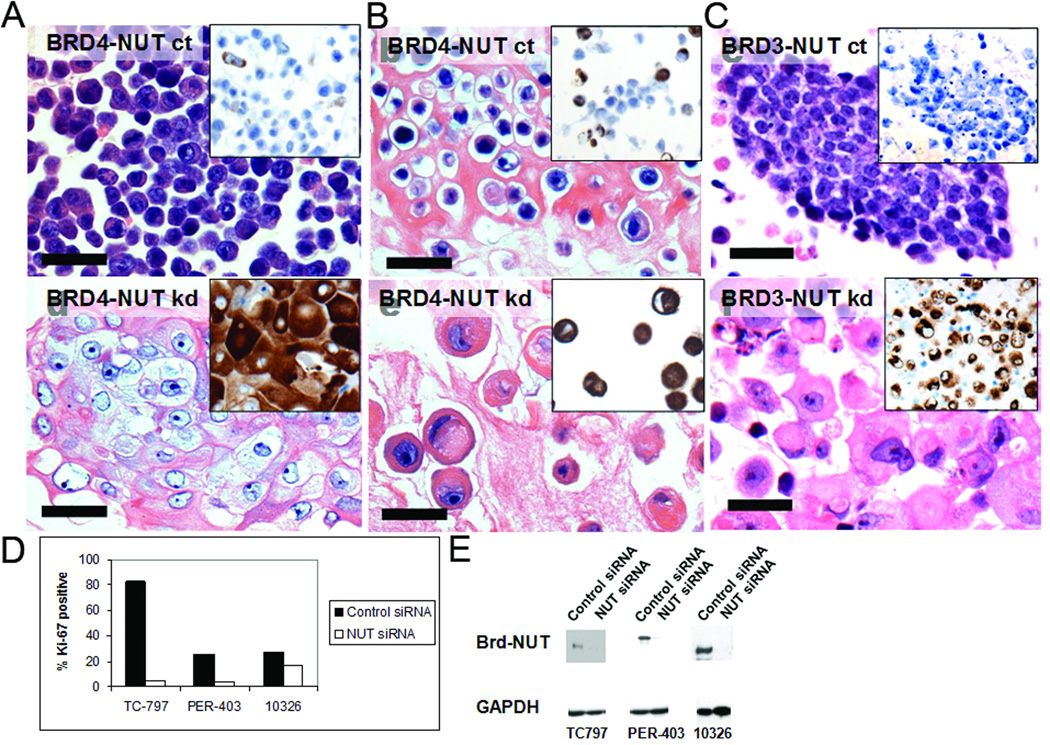
Involvement of BRD4 and possibly NUT in chromatin regulation suggested that BRD4-NUT might somehow modify chromatin so as to prevent the expression of genes needed for epithelial differentiation. Indeed, as with PML-RARα in acute promyelocytic leukemia, abrogation of BRD4-NUT or BRD3-NUT activity in NMC cell lines through siRNA-mediated knockdown results in induction of squamous differentiation followed by growth arrest (Fig. 2, [20]). Long term studies (3 weeks) have indeed found that the differentiation is terminal and irreversible. The findings suggest that BRD4-NUT is responsible for blocking differentiation and maintaining the cells in a perpetual state of proliferation, and that BRD4-NUT is potentially a powerful therapeutic target.
FIG 2.
Knockdown of BRD4-NUT results in squamous differentiation and growth arrest (from [20]). (A-C) Control siRNA (ct) has no effect on the BRD4-NUT-expressing cell lines TC797 and PER-403 [10], or the BRD3-NUT-expressing cell line 10326, resp. (hematoxylin and eosin stain, 400x, 96h post-transfection). Morphologic changes induced by NUT siRNA-induced knockdown (kd) in TC797, PER-403, and 10326 cells (hematoxylin and eosin stain, 400x, 96h post-transfection). (A-C, insets) BRD-NUT knockdown induces keratin expression, a biomarker of epithelial differentiation, as evidenced by increased staining for pan-keratin monoclonal antibody (clone MNF116, DAKO). (D) Knockdown of BRD-NUT decreases proliferation. Staining for Ki-67+ (a marker of cell cycle progression) was performed on sections of cell blocks prepared 96h post-siRNA transfection. Bars = 50 µm, A-C. (E) Effects of a _NUT_-specific siRNA on BRD-NUT. Protein extracts obtained 24h after siRNA transfection were analyzed on a Western blot stained with a polyclonal NUT antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Vargas SO, French CA, Faul PN, Fletcher JA, Davis IJ, Dal Cin P, Perez-Atayde AR. Upper respiratory tract carcinoma with chromosomal translocation 15;19: evidence for a distinct disease entity of young patients with a rapidly fatal course. Cancer. 2001;92:1195–1203. doi: 10.1002/1097-0142(20010901)92:5<1195::aid-cncr1438>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Kuzume T, Kubonishi I, Takeuchi S, Takeuchi T, Iwata J, Sonobe H, Ohtsuki Y, Miyoshi I. Establishment and characterization of a thymic carcinoma cell line (Ty-82) carrying t(15;19)(q15;p13) chromosome abnormality. Int J Cancer. 1992;50:259–264. doi: 10.1002/ijc.2910500216. [DOI] [PubMed] [Google Scholar]
- 3.French CA, Miyoshi I, Aster JC, et al. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t (15;19) Am J Pathol. 2001;159(6):1987–1992. doi: 10.1016/S0002-9440(10)63049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–307. [PubMed] [Google Scholar]
- 5.French CA, Kutok JL, Faquin WC, Toretsky JA, Antonescu CR, Griffin CA, Nose V, Vargas SO, Moschovi M, Tzortzatou-Stathopoulou F, Miyoshi I, Perez-Atayde AR, Aster JC, Fletcher JA. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol. 2004;22:4135–4139. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 6.Stelow EB, Bellizzi AM, Taneja K, Mills SE, Legallo RD, Kutok JL, Aster JC, French CA. NUT rearrangement in undifferentiated carcinomas of the upper aerodigestive tract. The American journal of surgical pathology. 2008;32:828–834. doi: 10.1097/PAS.0b013e31815a3900. [DOI] [PubMed] [Google Scholar]
- 7.French CA. Molecular pathology of NUT midline carcinomas. J Clin Pathol. 2008 doi: 10.1136/jcp.2007.052902. [DOI] [PubMed] [Google Scholar]
- 8.Lee AC, Kwong YI, Fu KH, Chan GC, Ma L, Lau YL. Disseminated mediastinal carcinoma with chromosomal translocation (15;19). A distinctive clinicopathologic syndrome. Cancer. 1993;72:2273–2276. doi: 10.1002/1097-0142(19931001)72:7<2273::aid-cncr2820720735>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 9.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, Roberts JR, Carbone DP. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. Journal of the National Cancer Institute. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 10.Kees UR, Mulcahy MT, Willoughby ML. Intrathoracic carcinoma in an 11-year-old girl showing a translocation t(15;19) The American journal of pediatric hematology/oncology. 1991;13:459–464. doi: 10.1097/00043426-199124000-00011. [DOI] [PubMed] [Google Scholar]
- 11.French CA, Miyoshi I, Aster JC, Kubonishi I, Kroll TG, Dal Cin P, Vargas SO, Perez-Atayde AR, Fletcher JA. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19) The American journal of pathology. 2001;159:1987–1992. doi: 10.1016/S0002-9440(10)63049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toretsky JA, Jenson J, Sun CC, Eskenazi AE, Campbell A, Hunger SP, Caires A, Frantz C, Hill JL, Stamberg J. Translocation (11;15;19): a highly specific chromosome rearrangement associated with poorly differentiated thymic carcinoma in young patients. Am J Clin Oncol. 2003;26:300–306. doi: 10.1097/01.COC.0000020960.98562.84. [DOI] [PubMed] [Google Scholar]
- 13.French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, Fletcher JA, Aster JC. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2007 doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 14.Kubonishi I, Takehara N, Iwata J, Sonobe H, Ohtsuki Y, Abe T, Miyoshi I. Novel t(15;19)(q15;p13) chromosome abnormality in a thymic carcinoma. Cancer research. 1991;51:3327–3328. [PubMed] [Google Scholar]
- 15.Engleson J, Soller M, Panagopoulos I, Dahlen A, Dictor M, Jerkeman M. Midline carcinoma with t(15;19) and BRD4-NUT fusion oncogene in a 30-year-old female with response to docetaxel and radiotherapy. BMC cancer. 2006;6:69. doi: 10.1186/1471-2407-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertens F, Wiebe T, Adlercreutz C, Mandahl N, French CA. Successful treatment of a child with t(15;19)-positive tumor. Pediatr Blood Cancer. 2007;49:1015–1017. doi: 10.1002/pbc.20755. [DOI] [PubMed] [Google Scholar]
- 17.den Bakker MD, Beverloo HB, van den Heuvel-Eibrink MM, Meeuwis CA, Tan ML, Johnson LA, French CA, van Leenders GJLH. NUT midline carcinoma of the parotid gland with mesenchymal differentiation. American Journal of Surgical Pathology. 2009 doi: 10.1097/PAS.0b013e3181abe120. in press. [DOI] [PubMed] [Google Scholar]
- 18.Ziai J, French CA, Zambrano E. NUT Gene Rearrangement in a Poorly-differentiated Carcinoma of the Submandibular Gland. Head and neck pathology. doi: 10.1007/s12105-010-0174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haack H, Johnson LA, Fry CJ, Crosby K, Polakiewicz RD, Stelow EB, Hong SM, Schwartz BE, Cameron MJ, Rubin MA, Chang MC, Aster JC, French CA. Diagnosis of NUT Midline Carcinoma Using a NUT-specific Monoclonal Antibody. The American journal of surgical pathology. 2009 doi: 10.1097/PAS.0b013e318198d666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, Fletcher JA, Aster JC. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 21.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 24.Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. The Journal of biological chemistry. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]