Interferon-Stimulated Gene 15 and the Protein ISGylation System (original) (raw)
Abstract
Interferon-stimulated gene 15 (ISG15) is one of the most upregulated genes upon Type I interferon treatment or pathogen infection. Its 17 kDa protein product, ISG15, was the first ubiquitin-like modifier identified, and is similar to a ubiquitin linear dimer. As ISG15 modifies proteins in a similar manner to ubiquitylation, protein conjugation by ISG15 is termed ISGylation. Some of the primary enzymes that promote ISGylation are also involved in ubiquitin conjugation. The process to remove ISG15 from its conjugated proteins, termed de-ISGylation, is performed by a cellular ISG15-specific protease, ubiquitin-specific proteases with molecular mass 43 kDa (UBP43)/ubiquitin-specific proteases 18. Relative to ubiquitin, the biological function of ISG15 is still poorly understood, but ISG15 appears to play important roles in various biological and cellular functions. Therefore, there is growing interest in ISG15, as the study of free ISG15 and functional consequences of ISGylation/de-ISGylation may identify useful therapeutic targets. This review highlights recent discoveries and remaining questions important to understanding the biological functions of ISG15.
Discovery of Interferon-Stimulated Gene 15
Interferon-stimulated gene 15 (ISG15) was first described in 1979 as a ∼15 kDa protein translated in vitro from interferon (IFN)-stimulated murine tumor cell RNA (Farrell and others 1979). The detection of this protein was next reported in 1984 from human and bovine cell lines treated with IFN, especially the Type I IFNs, IFN-α and IFN-β (Korant and others 1984). The first human sequence was identified in 1986 by cloning cDNA from human B lymphoblastic Daudi cells (Blomstrom and others 1986), although the original sequence included an extra nucleotide in error. When ISG15 was re-cloned in 1987, it correctly contained a full-length coding sequence predicting a protein of 165 amino acids and a molecular weight of 17,890 Da (Reich and others 1987). The full-length human ISG15 protein is actually a precursor to a shorter, more mature form generated by removing 8 C-terminal amino acids (Knight and others 1988). The mature form of ISG15 has a calculated molecular weight of 17,145 Da, lacks an N-terminal methionine, and has a C-terminus ending with the amino acid sequence Leu Arg Leu Arg Gly Gly (LRLRGG) (Knight and others 1988), which is the same 6 amino-acid sequence as mature ubiquitin. As late as 1987, ISG15 was still identified as a ubiquitin cross-reactive protein due to its cross-reactivity with certain anti-ubiquitin antibodies (Haas and others 1987), but ubiquitin cross-reactive protein was identical to the originally identified 17 kDa IFN-inducible protein. The name ISG15 was finally coined in the same year (Reich and others 1987).
ISG15 Sequence Homology
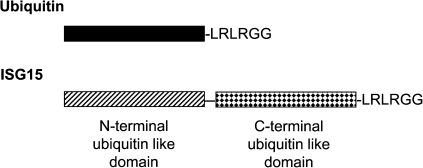
ISG15 contains 2 ubiquitin-like domains, making it a linear dimer of a ubiquitin-like protein (Fig. 1). ISG15 is identified in various mammals, including human, monkey, panda, horse, cow, sheep, pig, dog, cat, rabbit, rat, mouse, and opossum. Compared with ubiquitin, which has virtually 100% cross-species conservation, ISG15 protein has relatively low cross-species conservation, ranging from a high of 98% (chimpanzee to human) to a low of 42% (opossum to human) in mammals. ISG15 has also been cloned from many teleost species, including goldfish, zebrafish, catfish, pufferfish, salmon, cod, rockfish, and flounder, with a much lower amino acid similarity to mammals (∼30% with human ISG15). Further, although ubiquitin and many ubiquitin-like proteins, such as SUMO1 and NEDD8, are in all species of eukaryotic cells, ISG15 is only identified in vertebrates. Together, the relatively low cross-species conservation and absence in many eukaryotic species indicate that ISG15 is not an essential housekeeping gene, leaving more room for its diversification during evolution. Although all reported ISG15 proteins have the highly conserved C-terminal LRLRGG sequence, ISG15 in some species, such as fish, cow, and sheep, lack the short extension following the LRLRGG (Ritchie and Zhang 2004).
FIG. 1.
A domain diagram of ubiquitin and ISG15. ISG15, interferon-stimulated gene 15; LRLRGG, Leu Arg Leu Arg Gly Gly.
Characterization of ISG15 Gene Expression and Biological Function of Free ISG15
ISG15 induction is primarily triggered by Type I IFNs. The promoter of ISG15 contains 2 IFN-stimulated response elements (ISREs). The ISG15 promoter and the ISG54 promoter were initially used to identify the ISRE sequence and to purify transcription factors binding to this site, including both STAT1 and STAT2 (Reich and others 1987; Fu and others 1992). Therefore, ISG15 has a special role in the history of describing the JAK-STAT signaling pathway. Since Type III IFNs essentially use the same signaling pathway as Type I IFNs (Kotenko and Langer 2004), they also activate ISG15 production. However, the intensity of ISG15 induction is weaker by Type III IFNs than that by Type I IFNs (Burkart and Zhang, unpublished data). Further, since IFN response factors (IRFs) recognize a DNA sequence similar to the ISRE, they should also activate ISG15 expression. ISG15 expression is further regulated by PU.1, a member of the Ets family of transcription factors. PU.1 is B cell and myeloid cell-specific and plays an important role in blood cell differentiation and function (Tenen and others 1997). The promoter region of ISG15 contains a PU.1 binding site, which overlaps with the ISRE sequence. PU.1 synergistically induces expression of ISG15 with IRF4 or IRF8 (Meraro and others 2002), suggesting that PU.1 contributes to higher expression of ISG15 and protein ISGylation observed in myeloid cells as compared to nonhematopoietic cells.
Similar to other ISGs, ISG15 expression is upregulated after cell stress, especially those induced by bacterial and viral infections. These stresses activate transcription factors in IFN signaling, mainly IRF3 and ISGF3 (Nicholl and others 2000; Mossman and others 2001), which in turn upregulate expression of ISG15. Both external insults (gamma irradiation, anti cancer drugs, or viral infection) and internal insults (diseases and aging) can trigger ISG15 expression. In Type I IFN receptor R1 knockout mice and cells, ISG15 production dramatically decreases upon treatment with Gram-negative bacteria lipopolysaccharide (LPS) or viral infections (Kim and others 2005; Lenschow and others 2005), suggesting that stress induces Type I IFN secretion and triggers ISG15 expression. The remaining weak and short-term induction of ISG15 expression in the absence of Type I IFN signaling is probably due to the direct activation of IRFs. Nitrosylation of ISG15 prevents its dimerization and causes ISG15 activation (Okumura and others 2008b). Nitrosylation may occur under oxidative stress, which provides another mechanism of ISG15 activation. ISG15 is upregulated during telomere length shortening in a Type I IFN-independent manner in human fibroblasts (Lou and others 2009), but the molecular mechanism of this regulation is unclear.
Free ISG15 is also secreted from cells and detectable in both blood and urine as a cytokine and chemokine. ISG15 stimulates the production of Type II IFN, IFNγ, from CD3+ T cells to enhance the proliferation and cytotoxicity of natural killer cells (D'Cunha and others 1996). ISG15 is also a neutrophil chemotactic factor, as shown in _Plasmodium yoelii_–infected red blood cells (Owhashi and others 2003). However, cytokine or chemokine-related defects in ISG15 knockout mice have not been identified (Osiak and others 2005).
Free ISG15 also interacts with ubiquitin E3 Nedd4 and blocks Nedd4 activity in ubiquitylating Ebola virus VP40 protein (Malakhova and Zhang 2008; Okumura and others 2008a). The decreased ubiquitylation of VP40 significantly reduces the efficient release of VP40 virus-like particles from cells. Free ISG15 blocks the interaction of Nedd4 with ubiquitin E2 proteins, thus preventing further ubiquitin transfer from E2 to E3 (Malakhova and Zhang 2008; Okumura and others 2008a). Since ISG15 expression is dramatically increased upon Type I IFN stimulation, this novel mechanism of free ISG15 action in blocking protein–protein interaction provides a unique angle to study the role of ISG15 in innate immune responses upon various cellular stresses.
Structure of ISG15 Protein
ISG15 contains 2 ubiquitin-like domains at both termini. The N-terminal and C-terminal domains share ∼33% and 32% homology with ubiquitin, respectively (Fig. 1). Further, they share conformational similarities to ubiquitin, as shown by the crystal structure at 2.4Å (Narasimhan and others 2005). ISG15 contains a central random coil hinge connecting 2 almost identical ubiquitin-like domains with a β-grasp fold of 5 β-sheets and 1 α-helix. The ubiquitin-like domains of ISG15 share a similar 3-dimensional structure with ubiquitin and the 2 ubiquitin-like modifiers, NEDD8 and SUMO1. The remarkable conservation between NEDD8 and the C-terminal domain of ISG15 enabled modeling of the binding of ISG15 within the adenylate active site of the ISG15-activating enzyme, [ubiquitin activating enzyme E1-like (UbE1L)] (Narasimhan and others 2005). The model predicts extensive contacts between the C-terminal domain of ISG15 and UbE1L and identifies 3 unique amino acid residues, Lys90, Trp123, and Phe149, for specificity of UbE1L. The model also suggests that the amphipathic N-terminal domain of ISG15 is solvent-exposed and has a different role than the C-terminal domain. Indeed, a later study showed that the N-terminal domain of ISG15 is required for the efficient E3 ligase-mediated transfer of ISG15 from the E2 enzyme UbCH8 to its substrates although it is dispensable in the activation and transthiolation steps (Chang and others 2008). The C-terminal domain of ISG15 is sufficient to link ISG15 to the E1 enzyme UbE1L and the E2 enzyme UbCH8. Thus, the 2 ubiquitin domains of ISG15 play different critical roles in ISGylation, which is unique among ubiquitin and ubiquitin-like modifiers.
Components of the ISGylation and de-ISGylation Systems
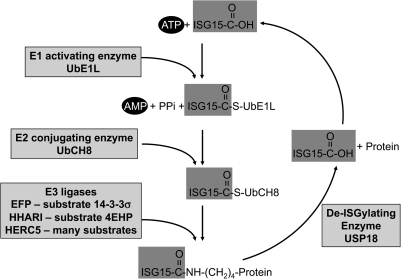
The modification of protein substrates with ISG15, termed ISGylation, is similar to ubiquitylation and uses a set of enzymes analogous to the ubiquitin modification system (Fig. 2). Protein ubiquitylation involves the coordinated activities of 3 modification enzymes: the activating or E1, the conjugating or E2, and the ligating or E3 enzymes (Pickart 2001). E1 proteins have conserved ATP-binding domains and active-site cysteine residues that are necessary for ubiquitin activation. The activated ubiquitin is then transferred to the active-site cysteine residue of an E2 enzyme. Finally, with the aid of an E3 ligase, the activated ubiquitin is transferred to a lysine residue of the substrate. Meanwhile, ubiquitin deconjugating enzymes (Dubs) facilitate the recycling of ubiquitin. For protein ISGylation, as shown in Fig. 2, UbE1L (aka UbA7) is a 112 kDa protein that has specific E1 activity (Yuan and Krug 2001). UbCH8 proteins from both human and mouse cells are the E2 enzyme for ISG15 (Kim and others 2004; Zhao and others 2004), although it may also function in ubiquitylation. Three cellular ISG15 E3 ligases have been identified so far, including estrogen-responsive finger protein (EFP) (aka TRIM25) (Zou and Zhang 2006), HERC5 (Dastur and others 2006), and human homolog of Drosophila ariadne (HHARI) (Okumura and others 2007). Finally, ubiquitin-specific proteases with molecular mass 43 kDa (UBP43)/ubiquitin-specific proteases 18 (USP18) is a cellular ISG15 deconjugating enzyme (Malakhov and others 2002) responsible for removing ISG15 from its conjugated proteins (Fig. 2). No ISG15 processing enzyme is known that removes the terminal region of ISG15 to give its mature form.
FIG. 2.
Protein ISGylation and de-ISGylaiton system. UbE1L catalyzes adenylation and then forms a thioester bond with the C-terminal end of ISG15. Activated ISG15 is transferred to the ISG15-conjugating enzyme, UbCH8. ISG15 and UbCH8 are also covalently linked via a thioester bond. With the help of ISG15 E3 ligase, the C-terminus of ISG15 is conjugated to the ɛ-amine group of lysine on the substrate. Ubiquitin-specific proteases 18 removes ISG15 from the substrate.
ISG15 activating enzyme UbE1L
UbE1L was originally cloned from a human pre-B cell library in 1993 (Kok and others 1993). The amino acid sequence of UbE1L shows a striking homology with ubiquitin E1, sharing 45% identity and a conserved cysteine residue at the active site (Kok and others 1993). UbE1L was first identified as an E1 enzyme for ISG15 in the search for influenza virus protein NS1B interacting proteins (Yuan and Krug 2001). Mice lacking the UbE1L gene fail to form ISG15 conjugates (Kim and others 2006), confirming that UbE1L is the bona fide E1 enzyme for ISG15.
Immunostaining of endogenous UbE1L in MG37, an EBV-transformed B-cell line, and exogenous UbE1L in transfected COS-7 cells showed that UbE1L is primarily a cytosolic protein, although both nuclear and cytoplasmic localizations occur in the transfected small cell lung cancer cell line, GLC-45 (McLaughlin and others 2000). Like ISG15, UbE1L expression is upregulated by IFN treatment (de Veer and others 2001), consistent with a conserved ISRE site at the upstream regulatory sequence of the UbE1L gene. However, UbE1L is constitutively expressed and shows weaker upregulation by IFN stimulation than ISG15 or other related ISG15 modification enzymes (Kim and others 2005).
ISG15 conjugating enzyme UbCH8
UbCH8 (aka UbE2L6), the E2 conjugating enzyme for ISG15, has been identified independently by 2 groups using differing approaches (Kim and others 2004; Zhao and others 2004). On the basis of the hypothesis that ISG15 would behave in the same manner as ubiquitin and form a thioester bond with an E2 enzyme, Zhao and others (2004) performed GST-ISG15 pull down experiments to identify proteins that would covalently link to ISG15 in protein extracts from IFN-treated cells. This approach allowed them to identify human UbCH8 as an ISG15 E2 enzyme. Kim and others began with the hypothesis that an E2 enzyme for ISG15 was potentially a member of the ubiquitin E2 family and that expression of an ISG15 conjugating enzyme would be IFN inducible, as both the E1 and deconjugating enzymes for ubiquitin and ISG15 share high homology and enzymes for ISG15 modification are IFN-inducible. The mouse E2 family members 1-8U, 9-27 (aka Leu13), and UbCH8 met these criteria and were tested for ISG15 E2 activity. UbCH8 was confirmed as an E2 enzyme using a co-transfection system to determine the relative levels of protein ISGylation (Kim and others 2004). Therefore, both approaches have identified UbCH8 as the ISG15 conjugating enzyme, although it also works as an E2 enzyme for ubiquitin. The UbCH8 promoter has both ISRE and PU.1 binding sites.
Among ubiquitin E2s, UbCH7 is most similar in primary sequence to UbCH8, but does not form a thioester bond with ISG15 in vitro (Zhao and others 2004) and does not enhance ISGylation in 293T cells (Kim and others 2004). Kinetic analysis indicated a strong preference for UbE1L to UbCH8 over UbCH7 (a 29-fold km) and a preference for ubiquitin E1 to UbCH7 over UbCH8 (a 36-fold km) (Durfee and others 2008). The N-terminal region of UbCH8 and a C-terminal domain of UbE1L play major roles in determining the specificity of the UbE1L–UbCH8 interaction. Further, since expression of both UbCH8 and ISG15 is strongly activated by IFN, ISG15 may be the major substrate of UbCH8 in vivo. The UbCH8 gene is located on human chromosome 11q12 and encodes a 16 kD protein.
ISG15 E3 ligases
E3 enzymes play primary roles in dictating substrate specificity. Mechanistically, ubiquitin E3 enzymes have been divided into 2 major groups—molecules containing domains homologous to the E6AP carboxy terminus (HECT) (Huibregtse and others 1995) and non-HECT E3 ligases. The non-HECT E3s contain a really interesting new gene (RING) finger domain or structurally related domains such as the U-box domain (Joazeiro and Weissman 2000; Hatakeyama and Nakayama 2003). HECT E3s accept ubiquitin from an E2 molecule and form a thioester adduct between HECT E3 and ubiquitin. Ubiquitin is transferred from the active-site cysteine of the HECT E3 ligase to a substrate via a covalent attachment to an ɛ-amino group of a lysine side chain (Huibregtse and others 1995). RING E3 molecules serve as docking proteins and bring together E2 molecules and substrates (Weissman 2001). RING domains contain 2 zinc ions coordinated by cysteines and histidines in a cross-brace structure. Unlike HECT E3s, RING E3s are noncatalytic.
Much progress has been made in recent years in the identification of ISG15 E3 ligases. The IFN-inducible ubiquitin E3 EFP (also known as TRIM25 for tripartite motif-containing 25) was identified as an ISG15 E3 ligase along with its substrate 14-3-3σ protein (Zou and Zhang 2006) using a similar strategy as Kim and others to identify ISG15 E2. In addition, the enzymatic activity of EFP is negatively regulated by auto-ISGylation at K117 (Zou and others 2007). Of E3s that function with the ISG15 E2 UbCH8 in protein ubiquitylation, HHARI is a specific E3 for ISGylation of 4EHP (Okumura and others 2007). Another E3 ligase for ISG15, HERC5, was identified among ubiquitin E3 ligases that are induced and ISGylated after IFN treatment (Dastur and others 2006). Reduction of endogenous HERC5 by small interfering RNAs blocks ISG15 conjugation to most target proteins, and overexpression of HERC5 produces robust ISGylation independent of IFN treatment, suggesting that HERC5 may not depend on its binding specificity toward any particular substrates although substrate specificity is a general feature E3 ligases. Further differing from RING finger E3 EFP and HHARI, HERC5 contains an HECT domain with a critical C994 residue for its enzymatic activity (Wong and others 2006). Mice do not have a direct HERC5 ortholog, but have HERC6 as an ISG15 E3 (Versteeg and others 2010), although human HERC6 is not an ISG15 E3. HERC5 and HERC6 are closely related HERC family proteins. In short, EFP and HHARI are substrate-specific E3s, whereas human HERC5 and murine HERC6 recognize ISG15, but do not have strong substrate specificity.
ISG15 deconjugating enzyme UBP43/USP18
USP18 was originally cloned and reported as UBP43 by our laboratory during the analysis of differentially expressed genes in AML1-ETO knock-in mice (Liu and others 1999). AML1-ETO is a fusion protein generated from an 8;21 chromosomal translocation and is associated with abnormal blood cell differentiation and proliferation. Heterozygous AML1-ETO knock-in mice die around E12.5 (Yergeau and others 1997). In AML1-ETO knock-in embryos, UBP43 is highly expressed in hematopoietic related organs, including both the yolk sac and fetal liver. UBP43 possesses cysteine and histidine boxes that are highly conserved and characteristic of the UBP family proteases (Liu and others 1999; Schwer and others 2000) involved in the removal of ubiquitin from conjugated proteins (Amerik and Hochstrasser 2004). Further, both human and mouse UBP43 mRNAs encode proteins with a calculated molecular weight of 43 kD, and are thus named UBP43. Recently, all human UBP family members were renamed as USP based on the order of their discovery. UBP43 is renamed as USP18 and we will use this new nomenclature from this point in the review.
Analysis using 125I-labelled ubiquitin and ubiquitin-like protein (ISG15, SUMO, and NEDD8) fusion peptides demonstrated that USP18 preferentially removes ISG15 from its conjugates (Malakhov and others 2002). The function of USP18 in removing ISG15 from its conjugates and the dependence of its enzyme activity on cysteine in the active site are further confirmed by in vivo studies (Malakhova and others 2006). The cloning of USP18 was also independently reported by 3 other groups in various species, and all confirmed the induction of USP18 by Type 1 IFN (Zhang and others 1999; Li and others 2000; Kang and others 2001), consistent with the finding of highly conserved ISREs in the UBP43 promoter (Malakhova and others 2002).
The Usp18 knockout mice show higher constitutive and IFN-inducible levels of ISG15 conjugation without obvious changes in protein ubiquitin conjugation (Ritchie and others 2002), indicating that USP18 is a bona fide ISG15 protease. Homozygous Usp18 knockout mice in an initial C57/B6 and 129 mixed strain showed a decreased lifespan mainly due to neurological abnormalities related to brain ependymal cell death leading to aqueduct stenosis and subsequent hydrocephalus (Ritchie and others 2002). With additional breeding within a C57/B6 and 129 mixed background, homozygous Usp18 knockout mice can have a nearly normal life span. After backcrossing heterozygous Usp18 knockout mice into a C57/B6 background, homozygous knockout mice die before E15.5. In the FVB/N background, a majority of _Usp18_−/− mice have a normal life span. Clearly, genetic background of these mice influences the phenotype of Usp18 knockout mice. Currently, _Usp18_−/− mice in the FVB/N background are available from Jackson Lab. _Usp18_-deficient cells are hypersensitive to IFN and LPS (Malakhova and others 2003; Kim and others 2005). Further, Usp18 knockout mice are resistant to viral and bacterial infections and are also resistant to BCR-ABL-induced leukemia development due to their enhanced responsiveness to Type I IFN (Ritchie and others 2004; Kim and others 2006, 2008a; Yan and others 2007). However, the enhanced sensitivity to IFN in _Usp18_-deficient cells is mainly independent of protein ISGylation and is due to the direct involvement of USP18 in IFN receptor-related JAK-STAT signal transduction (Malakhova and others 2006).
As the C-terminal regions of ISG15 and ubiquitin are quite similar, promiscuous de-ubiquitylating proteases may also recognize ISG15. For example, USP2, USP5, USP13, and USP14 are ISG15-reactive proteases (Catic and others 2007). However, the in vivo activity of these potential ISG15 deconjugating enzymes needs to be further clarified. In addition, several viral proteins are reported with de-ISGylation activity. A severe respiratory syndrome coronavirus papain-like protease uses both ISG15 and ubiquitin conjugates as its substrates (Lindner and others 2007). Similar proteases have been identified in other coronaviruses (Clementz and others 2010) and in 2 viral ovarian tumor-domain containing ubiquitin proteases (Frias-Staheli and others 2007).
Cellular Targets of the ISGylation System
Evidence for ISG15 protein conjugates was first reported in 1992 (Loeb and Haas 1992). After IFN treatment, free ISG15 levels increased in all 7 tested cell lines. However, protein ISGylation was only observed in 4 lines, A549, MG-63, U937, and Molt-4, and not the BCR/ABL+ leukemic cell line, K562, or 2 Burkitt's lymphoma cell lines, Daudi and Namalwa cells. Transfection of functional UbE1L into K562 cells restores ISGylation in these cells (Malakhova and others 2003). Similarly, Type I IFN stimulation induces ISGylation in HeLa, COS-7, RAW264.7, NIH3T3, 2fTGH, L929, A431, MCF7, and KT-1 cells, but not in Vero and 293T cells, although all cells showed increased levels of free ISG15 (Malakhova and others 2002; Liu and others 2003) (our unpublished data). The discrepancy between ISG15 induction and conjugation could result from a lack of functional ISG15 modification enzymes or the existence of factors that inhibit these enzymes. ISGylation is detected in liver, lung, heart, kidney, thymus, spleen, bone marrow, and brain after LPS or polyI-C injection in mice, suggesting that the molecular components required for ISGylation are widely distributed. Therefore, it is not surprising that ISGylation can be generally induced by diseases that trigger innate immune responses.
One key to understand biological function of ISGylation is identification of target proteins and specific ISG15 modification sites within these proteins (Table 1). Only 6 ISG15-conjugated proteins were reported in the literature for our last review in 2005 (Dao and Zhang 2005), Serpin 2a (Hamerman and others 2002), Jak1, Stat1, phospholipase C γ1, and extracellular regulated kinase 1/2 (Malakhov and others 2003), but this number has increased dramatically through the use of high-throughput approaches. Using immunoaffinity purification followed by mass spectrometry analysis, 76 candidate ISG15 conjugated proteins were identified from the human blood cell line, U937, and mouse embryonic fibroblasts treated with Type I IFN, 21 of which were found in both species (Giannakopoulos and others 2005). Eighteen of 19 proteins in this common group were confirmed in an ISGylation transfection system. The target proteins play important roles in translation, glycolysis, stress responses, and cell motility. Another study identified 158 putative ISGylated proteins and confirmed 8 proteins involved in various cellular functions, using NiNTA/FLAG double-affinity-purified protein samples from IFN-stimulated human HeLa cells co-transfected with HIS-FLAG double-tagged ISG15, UbE1L, and UbCH8 (Zhao and others 2005). Twenty-five proteins were identified by both studies in the putative ISGylated protein group, most of which play a role either in immunity, defense and stress responses, cell structure, and motility, or carbohydrate metabolism. A more recent high-throughput mass spectrometry study using FLAG-tagged, ISG15-expressing human A549 cells reported additional ISG15-conjugated proteins (Wong and others 2006). This target diversity suggests that protein ISGylation may be involved in regulating many cellular functions, as well as protein responses to Type I IFN production induced by pathogen infections and other stress conditions. A recent report revealed that HERC5 is associated with polyribosomes and that newly synthesized proteins are broadly targeted by ISG15 (Durfee and others 2010). This finding suggests the cotranslational ISGylation of both cellular and pathogen proteins with little specificity.
Table 1.
Interferon-Stimulated Gene 15 Target Proteins with Known Effect of Their ISGylation
| ISG15 targets | Function | ISGylation sites | Consequence of ISGylation | References |
|---|---|---|---|---|
| 4EHP | RNA 5' cap binding eIF4E family member, translation inhibitor | K134, K222 | Increase of its cap structure binding activity | Okumura and others (2007) |
| CHMP5 | A ESCRT-III protein | ND | Decrease of LIP5 and Vps4 interaction and block of retrovirus release | Pincetic and others (2010) |
| Cyclin D1 | Cell cycle activator | ND | Reduction of cellular cyclin D1 protein | Feng and others (2008) |
| EFP | Ubiquitin and ISG15 E3 ligase | K117 | Inhibition of its E3 ligase activity | Zou and others (2007) |
| Filamin B | Molecular scaffold and actin crosslinking | K2467 | Inactivation of JNK signaling pathway | Jeon and others (2009) |
| HPV16 L1 | Human papillomavirus capsid protein | ND | Reduction of infectivity of HPV16 pseudoviruses | Durfee and others (2010) |
| IRF3 | Interferon regulatory transcription factor | K193, K360, K366 | Increase of its activity due to reduced ubiquitination | Shi and others (2010) |
| NS1A | Influenza A viral nonstructural protein | K41 | Disruption of NS1A homodimer formation and nuclear transport | Zhao and others (2010); Tang and others (2010) |
| PML-RARα | Leukemia promoting fusion protein from chromosomal translocation | ND | Decrease of its stability | Shah and others (2008) |
| UbC13 | Ubiquitin E2 for K63-linked polyubiquitin chain | K92 | Disruption of thioester bond formation between ubiquitin and UbC13 to reduce E2 activity | Zou and others (2005); Takeuchi and Yokosawa (2005) |
| UbCH6 | Ubiquitin E2 for K63-linked polyubiquitin chain | K136 | Disruption of thioester bond formation between ubiquitin and UbCH6 to reduce E2 activity | Takeuchi and others (2005) |
Although there is no consensus modification site for protein ubiquitylation, a consensus modification site has been elucidated for the ubiquitin-like modifier, SUMO (Rodriguez and others 2001). Identifying the consensus motif for ISG15 conjugation may be an important step in elucidating the function of protein ISGylation. ISG15 modulation sites on the ɛ-amine group of lysine residues in a few target proteins have been reported, including K92 of UbC13 (Takeuchi and Yokosawa 2005; Zou and others 2005), K136 of UbCH6 (Takeuchi and others 2005), K117 of EFP (Zou and others 2007), K134/222 of 4EHP (Okumura and others 2007), K2467 of filamin B (Jeon and others 2009), K193/360/366 of IRF3 (Shi and others 2010), and K41 of the NS1 protein of influenza A virus (Zhao and others 2010). Comparison of target ISG15 sequences and mutagenesis studies may later reveal an ISGylation consensus motif.
Biological Function of Protein ISGylation
The conservation of ISG15 itself along with its conjugation and deconjugation enzymes in vertebrates and the tight regulation of ISG15 and its modification enzymes by Type I IFN stimulation strongly suggest the importance of this modification system in vertebrate biology. Nevertheless, a universal role of ISGylation has not yet been determined. The major question is whether there is a common fate for ISGylated proteins, similar to degradation of ubiquitylated proteins, or the fate varies according to the particular function of individual ISGylated proteins, as reported for conjugation by several other ubiquitin-like proteins. The answer to this question can only be obtained by comparing the role of ISGylation in different cellular systems.
Currently, there are 3 available mouse models to study the function of protein ISGylation: knockout mice of ISG15 (Osiak and others 2005), UbE1L (Kim and others 2006), and Usp18 (Ritchie and others 2002). ISG15 knockout mice lack both free ISG15 and ISG15 conjugates. UbE1L knockout mice lack ISGylated proteins but have higher than normal basal and inducible levels of free ISG15. Due to both the activity-dependent effect of Usp18 on ISG15 deconjugation and the activity-independent effect of Usp18 on Type I IFN signaling (Malakhova and others 2006), Usp18 knockout mice have much higher basal and inducible levels of ISG15 modified proteins and are hypersensitive to Type I IFNs and other reagents that stimulate Type I IFN production.
Protein ISGylation in innate immunity and anti-viral/bacterial infections
ISGylation putatively plays a role in innate immunity, as most components of the ISG15 modification system, including ISG15 and all its conjugating and deconjugating enzymes, are upregulated by type I IFN, and because many ISGylated proteins are involved in stress responses. For example, ISG15 expression from the recombinant chimeric Sindbis-virus resulted in resistance to Sindbis virus-induced lethality in IFNAR1 knockout mice (Lenschow and others 2005). Importantly, the C-terminal LRLRGG motif, which is required for ISG15 conjugation to other proteins, is necessary for resistance to Sinbis virus. Using ISG15 knockout mice, the same group later demonstrated that ISG15 is important for resistance to infections by influenza A and B viruses, herpes simplex virus type 1, murine gamma herpesvirus 68, and Sindbis virus (Lenschow and others 2007). Further, UbE1L knockout mice that lack protein ISGylation but express a high level of free ISG15 also exhibited a substantially increased susceptibility to influenza B virus infection (Lai and others 2009). However, wild-type, ISG15 knockout, and UbE1L knockout mice exhibited similar susceptibility to vesicular stomatitis virus and lymphocytic choriomeningitis virus infection (Osiak and others 2005; Kim and others 2006). Taken together, protein ISGylation plays important roles in defending against infections by select groups of viruses. Future work would be useful to characterize the basis of such selection.
Via either overexpression of exogenous ISG15 or decreasing endogenous ISG15 expression using an siRNA in cultured cells, all experimental results indicate that ISG15 inhibits infection and production of various viruses, such as HIV (Kunzi and Pitha 1996), Japanese encephalitis virus (Hsiao and others 2010), influenza A virus (Hsiang and others 2009), and vaccinia virus missing its early E3 protein (Guerra and others 2008). However, in the case of hepatitis C virus (HCV), ISG15 and ISGylation showed different effects on virus replication. Knockdown of ISG15 in HCV replicon-positive Huh-7 cells resulted in increased phenotypic sensitivity to IFN treatment that inhibited viral RNA replication (Chua and others 2009). A similar study using both knockdown and activation of the ISGylation system demonstrated that protein ISGylation promoted HCV production (Chen and others 2010b). This finding was also supported by the observation that increased ISG15 proteins in HCV-infected hepatocytes was associated with nonresponsiveness of patients to IFN therapy (Chen and others 2010a). HCV might therefore exploit ISG15 for host immune evasion. Higher levels of ISG15 are detected in Kupffer cells in IFN responders. Similarly, ISG15 modification of a key regulator of the IFN response, retinoic acid-inducible gene I (RIG-I), reduces total RIG-I levels and negatively regulates RIG-I function in IFNβ production and cellular IFN responses (Kim and others 2008b). Therefore, protein ISGylation may have multiple roles in innate immune responses depending on cell types or pathogens.
The role of ISGylation in innate immune responses is beginning to emerge. ISG15 enhances IRF3 activity by ISGylation of IRF3 to prevent its interaction with Pin1 for ubiquitylation and degradation (Shi and others 2010). ISG15 expression blocked retroviral budding (Pincetic and others 2010) by decreasing the interaction between vacuolar protein sorting 4 (Vps4) and its coactivator Lyst-interacting protein 5 (LIP5) due to ISG15 conjugation to the endosomal sorting complex required for transport complex III protein, CHMP5. A more direct molecular mechanism for the anti-viral activities of ISG15 was discovered in a study of the influenza A viral protein NS1 (NS1A). Influenza A virus blocks cellular ISG15 production upon infection. In contrast, influenza B virus triggered high levels of ISG15 synthesis, but the viral NS1B protein interacts with ISG15 to block its interaction with UbE1L and stop activation of ISGylation (Yuan and Krug 2001). Thus, ISG15 conjugation may be detrimental to influenza infection. Multiple lysines of the NS1A protein could be modified by ISG15, with K41 in the N-terminal domain of this protein acting as the main modification site (Tang and others 2010; Zhao and others 2010). This domain is responsible for binding to double-stranded RNA and NS1A nuclear localization via interaction with a cellular protein importin-α. The nuclear localization is essential for influenza A virus replication and blocking cellular RNA processing. ISGylation at K41 does not affect RNA binding but disrupts NS1A interactions with importin-α and nuclear transport (Zhao and others 2010). The ISGylated NS1A also failed to form homodimers, a required step for normal function, and HERC5 directly interacts with and ISGylates NS1A (Tang and others 2010). Consequently, ISGylation of NS1A inhibits viral infection. Further, ISGylation of HPV16 L1 protein is reported to have a dominant negative effect on infectivity of HPV pseudovirus (Durfee and others 2010).
Protein ISGylation in relation to proteasome function in protein turnover
Given the role for ubiquitin in protein degradation and the similarity of ISG15 and ubiquitin modifications, ISG15 may influence protein turnover. The involvement of ubiquitin-like modifiers in protein degradation has been observed in other cases. For example, proteins modified by NEDD8 were eventually degraded by the proteasome, which was facilitated by the adaptor protein NUB1 (NEDD8 ultimate buster 1) (Kamitani and others 2001). This protein degradation does not act directly through NEDD8 conjugation but rather indirectly through the recruitment of a ubiquitin-conjugating protein complex. Protein modification by another ubiquitin-like modifier, SUMO, also counteracts and facilitates protein ubiquitylation for degradation (Geoffroy and Hay 2009). Further, modification by the ubiquitin-like modifier FAT10 promotes proteasome-mediated degradation of its conjugates (Hipp and others 2005).
Proteasome inhibitors, such as MG132 or lactacystin, increase levels of ISG15 conjugates (Liu and others 2003). However, unlike ubiquitin conjugates, this increase could be blocked by ATP depleting reagents. Thus, these increased ISG15 conjugate levels require de novo ISG15 conjugation, indicating ISG15 does not mark proteins for degradation through the proteasome pathway, otherwise proteasome inhibitors should increase ISG15 conjugates even in the presence of ATP depleting reagents (Liu and others 2003). We observed that these proteasome inhibitors did not affect the levels of protein ISGylation in the absence or presence of IFN stimulation (Malakhov and others 2003), although they did increase levels of ubiquitylated proteins. These results suggest differential cell-specific responses to proteasome inhibitor-induced stress. Regardless of this discrepancy, both groups concluded that protein ISGylation does not target proteins for degradation through the proteasomal pathway. However, ISGylation is still possible to influence cellular protein degradation.
One mechanism for ISG15 to regulate proteasome activity is through the ISGylation of modifying enzymes involved in ubiquitylation. ISGylation of an ubiquitin conjugating enzyme, UbC13, at K92 disrupts its ability to form a thioester bond with ubiquitin and blocks its ubiquitin-conjugating activity (Takeuchi and Yokosawa 2005; Zou and others 2005). K92 of UbC13 is the first ISG15 modulation site identified via mass spectrometry analysis (Zou and others 2005). Similarly, the activity of other ubiquitin E2 enzymes, UbCH6 and UbCH8, were suppressed by ISGylation, and K136 of UbCH6 is an ISG15 modulation site (Takeuchi and others 2005). ISG15 modification of EFP E3 ligase also inhibited its enzyme activity for ISGylation of 14-3-3σ (Zou and others 2007). Specific knockdown of ISG15 or its E2 conjugating enzyme, UbCH8, by small interfering RNA also increased the level of polyubiquitylated proteins in tumor cells that normally expressed an increased level of ISG15 (Desai and others 2006). Together, these studies suggest that ISGylation may negatively regulate ubiquitination and indirectly affect ubiquitin protein targets. However, general protein ubiquitylation patterns in ISG15 activating enzyme UbE1L+/+ and UbE1L−/− cells are similar even after IFN stimulation (Kim and others 2006), indicating that protein ISGylation is not tightly linked to protein ubiquitylation.
Protein ISGylation in tumorigenesis
Studies of ISG15 expression in primary tumor tissues and cell lines indicate a possible link between ISG15 and tumorigenesis. High levels of ISG15 and its conjugates occur in many types of primary tumors, including bladder, prostate, oral, and breast cancers (Andersen and others 2006; Bektas and others 2008; Chi and others 2009; Satake and others 2010). Such enhanced ISG15 expression and protein ISGylation are most likely triggered by increased IFN release when cancer is present. However, considering a recent report that telomere shortening may trigger ISG15 expression in an IFN-independent manner, more studies will facilitate our understanding of induction of ISG15 in cancer. Robust expression of ISG15 in cancer tissues may serve as a useful biomarker for cancer and for chemotherapeutic drug sensitivity studies (Bektas and others 2008; Desai and others 2008; Weichselbaum and others 2008).
The possible connection of ISGylation to cancer was initiated by the discovery of the UbE1L gene locus on chromosome 3p21. Loss of allelic heterozygosity in chromosome 3p21 is observed in 70%–80% of nonsmall-cell lung cancers and 90%–100% of small cell lung cancers (Kok and others 1993). Therefore, UbE1L is a candidate tumor suppressor gene at this locus. Two lung cancer mouse models, K-rasLA2 and p53-deficient mice, were used to address the role of protein ISGylation in cancer development by breeding these mice with UbE1L knockout mice. Besides modulating the tumor spectrum, no significant differences in lung cancer development were observed in UbE1L+/+ and _UbE1L_−/− mice (Yin and others 2009a, 2009b), suggesting that UbE1L did not play a critical tumor suppressor role in K-rasG12D and p53 deficiency-induced lung cancer. Additional work with other mouse cancer models should provide insight into the role of protein ISGylation in cancer development.
Several studies using in vitro cell lines and human cancer samples begin to reveal different roles for ISGylation in tumorigenesis. ISGylation might negatively regulate ubiquitylation in tumor cells that express increased levels of ISG15 (Desai and others 2006). Retinoic acid treatment activates Type I IFN-mediated gene expression, including both UbE1L and ISG15, and leads to consequent protein ISGylation (Kitareewan and others 2002; Dao and others 2006). Activation of UbE1L expression in several cancer cell lines promotes cyclin D1 and PML-RARα ISGylation and decreases their overall protein levels, which was associated with decreased cell growth (Feng and others 2008; Shah and others 2008). In contrast, increased expression of ISG15, UbE1L, and several other ISGylation-related enzymes were detected in androgen receptor-positive prostate cancer samples (Kiessling and others 2009). Overexpression of UbE1L enhanced levels of androgen receptor mRNA and protein in an ISG15-dependent manner. This implies that protein ISGylation promotes expression of androgen receptor. Further, the same cell line with increased UbE1L expression conferred via stable transfection showed increased proliferation upon androgen stimulation, suggesting that UbE1L and protein ISGylation promote prostate cancer development (Kiessling and others 2009). Together, these studies indicate that protein ISGylation may have opposing effects on tumorigenesis in different cell types and during different stages of cancer development. This is an important topic and warrants further investigation.
Knocking down ISG15 or UbCH8 in breast cancer ZR-751 cells decreases their sensitivity to chemotherapeutic drug camptothecins (Desai and others 2008). ISG15 levels are low in several tumor cell lines resistant to chemotherapeutic drug camptothecins. Consistent with this, many lung cancers, solid tumors, and an IFN-insensitive leukemia cell line lack functional UbE1L (Kok and others 1993; Malakhova and others 2003). The BCR-ABL transformation of hematopoietic cells suppresses IFN-inducible transcription of ISG15 (Katsoulidis and others 2008) and Usp18-deficient mice showed increased resistance to oncogenic transformation by BCR-ABL (Yan and others 2007). In addition, Type I IFNs enhance the effect of the chemotherapeutic drug 5-fluorouracil on cell lines derived from esophageal cancer with high levels of ISG15 (Matsumura and others 2005). However, the opposite result was observed for chemoresistance to gemcitabine, a standard chemotherapeutic agent for pancreatic cancer (Ina and others 2010), where knocking down ISG15 in pancreatic cancer cell lines reversed gemcitabine resistance. It is currently unclear whether this discrepancy is due to differences among chemotherapeutic drugs or cancer cell types.
Basal levels of cellular ISGylation are very low. UbE1L knockout mice are healthy and with similar peripheral and bone marrow blood cells as wild-type mice (Kim and others 2006; Cong and others 2010). Bone marrow transplantation induces transient Type I INF expression and protein ISGylation within 10 days of the procedure, and UbE1L-deficient blood progenitor cells showed a delayed proliferative response, especially at the G2/M phase of cell cycle (Cong and others 2010). ISG15 also modified filamin B, an important molecular scaffold of the cytoskeleton, and other cellular protein complexes, such as RAC1-MEKK1/MKK4/JNK (Jeon and others 2009). Blocking ISGylation of filamin B inhibited the dissociation of signaling molecules and resulted in persistent JNK activation and JNK-mediated apoptosis. A gain of function of protein ISGylation is reported in ISG15 modification of 4EHP. 4EHP is an mRNA 5′ cap structure-binding protein and suppresses translation by competing with eIF4E for binding to the cap structure. ISGylated 4EHP had higher cap structure binding, which may influence translational control of selected mRNA targets during immune responses (Okumura and others 2007).
Concluding Remarks
ISG15 was the first identified ubiquitin-like protein and modifier. Its presence in only vertebrates, relatively low cross-species sequence homology, and strong induction upon signal stimulation indicate that ISG15 is not essential for basic cellular function. Analyses of ISG15 and UbE1L knockout mice clearly demonstrate this point. ISG15 itself as well as most enzymes involved in protein ISGylation and de-ISGylation are regulated by Type I IFN stimulation. Therefore, ISG15 and protein ISGylation is a tightly controlled and energy-consuming process, suggesting their important and specialized functions in vertebrate animals. Since our last review in 2005 (Dao and Zhang 2005), we have identified nearly 200 new reports in PubMed with ISG15 as a key word, indicating vigorous research in this field. Indeed, revealing additional enzymes in the ISGylation process, identifying and confirming many ISG15 target proteins, and establishing ISG15 and UbE1L knockout mouse models in the last 5 years have greatly enriched our knowledge in the ISG15 field. Exciting reports are emerging about the biological function of ISG15 and protein ISGylation. For free ISG15, we are looking forward to having insightful information regarding its secretion pathway as a cytokine and chemokine, its cellular receptors, and pathways in signal transduction. Further, considering recent reports related to linear and K63-linked polyubiquitin in various immune responses, it would be very interesting to examine the parallel functions of ISG15. Regarding protein ISGylation, identifying ISG15 modification sites in more proteins will help us to understand the mechanism of modification by the ISGylation enzyme system, such as target recognition, a possible consensus modification site, and the consequence of such modification. To the biological function of protein ISGylation, novel gene knockout models, including ISG15, UbEL1, Usp18, and the expected HERC6 knockout mice, will continue to provide powerful tools for in vivo analysis. Future work will explore ISGylation in innate immune responses, the cross talk between the innate and adaptive immune systems, and its possible role during adaptive immune reactions. Further, we will better understand the link between protein ISGylation and maintaining protein stability, as well as the role of ISG15 and protein ISGylation in cancer development and as a biomarker for human diseases.
Acknowledgments
We thank Dr. Joseph Biggs for his critical reading of this article. The work in our laboratories related to this topic is supported by grants from the U.S. NIH (R21DE15129, R01GM66955, and R01HL091549).
Author Disclosure Statement
No competing financial interests exist.
References
- Amerik AY. Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695(1–3):189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Andersen JB. Aaboe M. Borden EC. Goloubeva OG. Hassel BA. Orntoft TF. Stage-associated overexpression of the ubiquitin-like protein, ISG15, in bladder cancer. Br J Cancer. 2006;94(10):1465–1471. doi: 10.1038/sj.bjc.6603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bektas N. Noetzel E. Veeck J. Press MF. Kristiansen G. Naami A. Hartmann A. Dimmler A. Beckmann MW. Knuchel R. Fasching PA. Dahl E. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Res. 2008;10(4):R58. doi: 10.1186/bcr2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomstrom DC. Fahey D. Kutny R. Korant BD. Knight E., Jr. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J Biol Chem. 1986;261(19):8811–8816. [PubMed] [Google Scholar]
- Catic A. Fiebiger E. Korbel GA. Blom D. Galardy PJ. Ploegh HL. Screen for ISG15-crossreactive deubiquitinases. PLoS One. 2007;2(7):e679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YG. Yan XZ. Xie YY. Gao XC. Song AX. Zhang DE. Hu HY. Different roles for two ubiquitin-like domains of ISG15 in protein modification. J Biol Chem. 2008;283(19):13370–13377. doi: 10.1074/jbc.M800162200. [DOI] [PubMed] [Google Scholar]
- Chen L. Borozan I. Sun J. Guindi M. Fischer S. Feld J. Anand N. Heathcote J. Edwards AM. McGilvray ID. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology. 2010a;138(3):1123–1133. doi: 10.1053/j.gastro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- Chen L. Sun J. Meng L. Heathcote J. Edwards AM. McGilvray ID. ISG15, a ubiquitin-like interferon-stimulated gene, promotes hepatitis C virus production in vitro: implications for chronic infection and response to treatment. J Gen Virol. 2010b;91(Pt 2):382–388. doi: 10.1099/vir.0.015388-0. [DOI] [PubMed] [Google Scholar]
- Chi LM. Lee CW. Chang KP. Hao SP. Lee HM. Liang Y. Hsueh C. Yu CJ. Lee IN. Chang YJ. Lee SY. Yeh YM. Chang YS. Chien KY. Yu JS. Enhanced interferon signaling pathway in oral cancer revealed by quantitative proteome analysis of microdissected specimens using 16O/18O labeling and integrated two-dimensional LC-ESI-MALDI tandem MS. Mol Cell Proteomics. 2009;8(7):1453–1474. doi: 10.1074/mcp.M800460-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PK. McCown MF. Rajyaguru S. Kular S. Varma R. Symons J. Chiu SS. Cammack N. Najera I. Modulation of alpha interferon anti-hepatitis C virus activity by ISG15. J Gen Virol. 2009;90(Pt 12):2929–2939. doi: 10.1099/vir.0.013128-0. [DOI] [PubMed] [Google Scholar]
- Clementz MA. Chen Z. Banach BS. Wang Y. Sun L. Ratia K. Baez-Santos YM. Wang J. Takayama J. Ghosh AK. Li K. Mesecar AD. Baker SC. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84(9):4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X. Yin X. Yan M. Zhang DE. Hematopoietic cells from Ube1l-deficient mice exhibit an impaired proliferation defect under the stress of bone marrow transplantation. Blood Cells Mol Dis. 2010;45(2):103–111. doi: 10.1016/j.bcmd.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao CT. Luo JK. Zhang DE. Retinoic acid-induced protein ISGylation is dependent on interferon signal transduction. Blood Cells Mol Dis. 2006;36(3):406–413. doi: 10.1016/j.bcmd.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Dao CT. Zhang DE. ISG15: a ubiquitin-like enigma. Front Biosci. 2005;10:2701–2722. doi: 10.2741/1730. [DOI] [PubMed] [Google Scholar]
- Dastur A. Beaudenon S. Kelley M. Krug RM. Huibregtse JM. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem. 2006;281(7):4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- D'Cunha J. Knight E., Jr. Haas AL. Truitt RL. Borden EC. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci USA. 1996;93(1):211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SD. Haas AL. Wood LM. Tsai YC. Pestka S. Rubin EH. Saleem A. Nur-E-Kamal Liu LF. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 2006;66(2):921–928. doi: 10.1158/0008-5472.CAN-05-1123. [DOI] [PubMed] [Google Scholar]
- Desai SD. Wood LM. Tsai YC. Hsieh TS. Marks JR. Scott GL. Giovanella BC. Liu LF. ISG15 as a novel tumor biomarker for drug sensitivity. Mol Cancer Ther. 2008;7(6):1430–1439. doi: 10.1158/1535-7163.MCT-07-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer MJ. Holko M. Frevel M. Walker E. Der S. Paranjape JM. Silverman RH. Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69(6):912–920. [PubMed] [Google Scholar]
- Durfee LA. Kelley ML. Huibregtse JM. The basis for selective E1-E2 interactions in the ISG15 conjugation system. J Biol Chem. 2008;283(35):23895–23902. doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee LA. Lyon N. Seo K. Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell. 2010;38(5):722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell PJ. Broeze RJ. Lengyel P. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature. 1979;279(5713):523–525. doi: 10.1038/279523a0. [DOI] [PubMed] [Google Scholar]
- Feng Q. Sekula D. Guo Y. Liu X. Black CC. Galimberti F. Shah SJ. Sempere LF. Memoli V. Andersen JB. Hassel BA. Dragnev K. Dmitrovsky E. UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol Cancer Ther. 2008;7(12):3780–3788. doi: 10.1158/1535-7163.MCT-08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N. Giannakopoulos NV. Kikkert M. Taylor SL. Bridgen A. Paragas J. Richt JA. Rowland RR. Schmaljohn CS. Lenschow DJ. Snijder EJ. Garcia-Sastre A. Virgin HW. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2(6):404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XY. Schindler C. Improta T. Aebersold R. Darnell JE., Jr. The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci USA. 1992;89(16):7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC. Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10(8):564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- Giannakopoulos NV. Luo JK. Papov V. Zou W. Lenschow DJ. Jacobs BS. Borden EC. Li J. Virgin HW. Zhang DE. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys Res Commun. 2005;336(2):496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- Guerra S. Caceres A. Knobeloch KP. Horak I. Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4(7):e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL. Ahrens P. Bright PM. Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262(23):11315–11323. [PubMed] [Google Scholar]
- Hamerman JA. Hayashi F. Schroeder LA. Gygi SP. Haas AL. Hampson L. Coughlin P. Aebersold R. Aderem A. Serpin 2a is induced in activated macrophages and conjugates to a ubiquitin homolog. J Immunol. 2002;168(5):2415–2423. doi: 10.4049/jimmunol.168.5.2415. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S. Nakayama KI. U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun. 2003;302(4):635–645. doi: 10.1016/s0006-291x(03)00245-6. [DOI] [PubMed] [Google Scholar]
- Hipp MS. Kalveram B. Raasi S. Groettrup M. Schmidtke G. FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol Cell Biol. 2005;25(9):3483–3491. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang TY. Zhao C. Krug RM. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J Virol. 2009;83(12):5971–5977. doi: 10.1128/JVI.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao NW. Chen JW. Yang TC. Orloff GM. Wu YY. Lai CH. Lan YC. Lin CW. ISG15 over-expression inhibits replication of the Japanese encephalitis virus in human medulloblastoma cells. Antiviral Res. 2010;85(3):504–511. doi: 10.1016/j.antiviral.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM. Scheffner M. Beaudenon S. Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92(7):2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ina S. Hirono S. Noda T. Yamaue H. Identifying molecular markers for chemosensitivity to gemcitabine in pancreatic cancer: increased expression of interferon-stimulated gene 15 kd is associated with intrinsic chemoresistance. Pancreas. 2010;39(4):473–485. doi: 10.1097/MPA.0b013e3181c0decc. [DOI] [PubMed] [Google Scholar]
- Jeon YJ. Choi JS. Lee JY. Yu KR. Kim SM. Ka SH. Oh KH. Kim KI. Zhang DE. Bang OS. Chung CH. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 2009;10(4):374–380. doi: 10.1038/embor.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA. Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102(5):549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Kamitani T. Kito K. Fukuda-Kamitani T. Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;276(49):46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- Kang D. Jiang H. Wu Q. Pestka S. Fisher PB. Cloning and characterization of human ubiquitin-processing protease-43 from terminally differentiated human melanoma cells using a rapid subtraction hybridization protocol RaSH. Gene. 2001;267(2):233–242. doi: 10.1016/s0378-1119(01)00384-5. [DOI] [PubMed] [Google Scholar]
- Katsoulidis E. Sassano A. Majchrzak-Kita B. Carayol N. Yoon P. Jordan A. Druker BJ. Fish EN. Platanias LC. Suppression of interferon (IFN)-inducible genes and IFN-mediated functional responses in BCR-ABL-expressing cells. J Biol Chem. 2008;283(16):10793–10803. doi: 10.1074/jbc.M706816200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling A. Hogrefe C. Erb S. Bobach C. Fuessel S. Wessjohann L. Seliger B. Expression, regulation and function of the ISGylation system in prostate cancer. Oncogene. 2009;28(28):2606–2620. doi: 10.1038/onc.2009.115. [DOI] [PubMed] [Google Scholar]
- Kim JH. Luo JK. Zhang DE. The level of hepatitis B virus replication is not affected by protein ISG15 modification but is reduced by inhibition of UBP43 (USP18) expression. J Immunol. 2008a;181(9):6467–6472. doi: 10.4049/jimmunol.181.9.6467. [DOI] [PubMed] [Google Scholar]
- Kim KI. Giannakopoulos NV. Virgin HW. Zhang DE. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol Cell Biol. 2004;24(21):9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KI. Malakhova OA. Hoebe K. Yan M. Beutler B. Zhang DE. Enhanced antibacterial potential in UBP43-deficient mice against Salmonella typhimurium infection by up-regulating type I IFN signaling. J Immunol. 2005;175(2):847–854. doi: 10.4049/jimmunol.175.2.847. [DOI] [PubMed] [Google Scholar]
- Kim KI. Yan M. Malakhova O. Luo JK. Shen MF. Zou W. de la Torre JC. Zhang DE. Ube1L and protein ISGylation are not essential for alpha/beta interferon signaling. Mol Cell Biol. 2006;26(2):472–479. doi: 10.1128/MCB.26.2.472-479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ. Hwang SY. Imaizumi T. Yoo JY. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008b;82(3):1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitareewan S. Pitha-Rowe I. Sekula D. Lowrey CH. Nemeth MJ. Golub TR. Freemantle SJ. Dmitrovsky E. UBE1L is a retinoid target that triggers PML/RARalpha degradation and apoptosis in acute promyelocytic leukemia. Proc Natl Acad Sci USA. 2002;99(6):3806–3811. doi: 10.1073/pnas.052011299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight E., Jr. Fahey D. Cordova B. Hillman M. Kutny R. Reich N. Blomstrom D. A 15-kDa interferon-induced protein is derived by COOH-terminal processing of a 17-kDa precursor. J Biol Chem. 1988;263(10):4520–4522. [PubMed] [Google Scholar]
- Kok K. Hofstra R. Pilz A. van den BA. Terpstra P. Buys CH. Carritt B. A gene in the chromosomal region 3p21 with greatly reduced expression in lung cancer is similar to the gene for ubiquitin-activating enzyme. Proc Natl Acad Sci USA. 1993;90(13):6071–6075. doi: 10.1073/pnas.90.13.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korant BD. Blomstrom DC. Jonak GJ. Knight E., Jr. Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J Biol Chem. 1984;259(23):14835–14839. [PubMed] [Google Scholar]
- Kotenko SV. Langer JA. Full house: 12 receptors for 27 cytokines. Int Immunopharmacol. 2004;4(5):593–608. doi: 10.1016/j.intimp.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Kunzi MS. Pitha PM. Role of interferon-stimulated gene ISG-15 in the interferon-omega-mediated inhibition of human immunodeficiency virus replication. J Interferon Cytokine Res. 1996;16(11):919–927. doi: 10.1089/jir.1996.16.919. [DOI] [PubMed] [Google Scholar]
- Lai C. Struckhoff JJ. Schneider J. Martinez-Sobrido L. Wolff T. Garcia-Sastre A. Zhang DE. Lenschow DJ. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol. 2009;83(2):1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ. Giannakopoulos NV. Gunn LJ. Johnston C. O'Guin AK. Schmidt RE. Levine B. Virgin HW. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol. 2005;79(22):13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ. Lai C. Frias-Staheli N. Giannakopoulos NV. Lutz A. Wolff T. Osiak A. Levine B. Schmidt RE. Garcia-Sastre A. Leib DA. Pekosz A. Knobeloch KP. Horak I. Virgin HW. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci USA. 2007;104(4):1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XL. Blackford JA. Judge CS. Liu M. Xiao W. Kalvakolanu DV. Hassel BA. RNase-L-dependent destabilization of interferon-induced mRNAs. A role for the 2-5A system in attenuation of the interferon response. J Biol Chem. 2000;275(12):8880–8888. doi: 10.1074/jbc.275.12.8880. [DOI] [PubMed] [Google Scholar]
- Lindner HA. Lytvyn V. Qi H. Lachance P. Ziomek E. Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys. 2007;466(1):8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LQ. Ilaria R., Jr. Kingsley PD. Iwama A. van Etten RA. Palis J. Zhang DE. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol Cell Biol. 1999;19(4):3029–3038. doi: 10.1128/mcb.19.4.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Li XL. Hassel BA. Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J Biol Chem. 2003;278(3):1594–1602. doi: 10.1074/jbc.M208123200. [DOI] [PubMed] [Google Scholar]
- Loeb KR. Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem. 1992;267(11):7806–7813. [PubMed] [Google Scholar]
- Lou Z. Wei J. Riethman H. Baur JA. Voglauer R. Shay JW. Wright WE. Telomere length regulates ISG15 expression in human cells. Aging. 2009;1(7):608–621. doi: 10.18632/aging.100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhov MP. Kim KI. Malakhova OA. Jacobs BS. Borden EC. Zhang DE. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J Biol Chem. 2003;278(19):16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- Malakhov MP. Malakhova OA. Kim KI. Ritchie KJ. Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277(12):9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- Malakhova O. Malakhov M. Hetherington C. Zhang DE. Lipopolysaccharide activates the expression of ISG15-specific protease UBP43 via interferon regulatory factor 3. J Biol Chem. 2002;277(17):14703–14711. doi: 10.1074/jbc.M111527200. [DOI] [PubMed] [Google Scholar]
- Malakhova OA. Kim KI. Luo JK. Zou W. Kumar KG. Fuchs SY. Shuai K. Zhang DE. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25(11):2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhova OA. Yan M. Malakhov MP. Ritchie KJ. Kim KI. Peterson LF. Shuai K. Zhang DE. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17(4):455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhova OA. Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283(14):8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y. Yashiro M. Ohira M. Tabuchi H. Hirakawa K. 5-Fluorouracil up-regulates interferon pathway gene expression in esophageal cancer cells. Anticancer Res. 2005;25(5):3271–3278. [PubMed] [Google Scholar]
- McLaughlin PM. Helfrich W. Kok K. Mulder M. Hu SW. Brinker MG. Ruiters MH. de Leij LF. Buys CH. The ubiquitin-activating enzyme E1-like protein in lung cancer cell lines. Int J Cancer. 2000;85(6):871–876. doi: 10.1002/(sici)1097-0215(20000315)85:6<871::aid-ijc22>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Meraro D. Gleit-Kielmanowicz M. Hauser H. Levi BZ. IFN-stimulated gene 15 is synergistically activated through interactions between the myelocyte/lymphocyte-specific transcription factors, PU.1, IFN regulatory factor-8/IFN consensus sequence binding protein, and IFN regulatory factor-4: characterization of a new subtype of IFN-stimulated response element. J Immunol. 2002;168(12):6224–6231. doi: 10.4049/jimmunol.168.12.6224. [DOI] [PubMed] [Google Scholar]
- Mossman KL. Macgregor PF. Rozmus JJ. Goryachev AB. Edwards AM. Smiley JR. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol. 2001;75(2):750–758. doi: 10.1128/JVI.75.2.750-758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan J. Wang M. Fu Z. Klein JM. Haas AL. Kim JJ. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J Biol Chem. 2005;280(29):27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- Nicholl MJ. Robinson LH. Preston CM. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J Gen Virol. 2000;81(Pt 9):2215–2218. doi: 10.1099/0022-1317-81-9-2215. [DOI] [PubMed] [Google Scholar]
- Okumura A. Pitha PM. Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci USA. 2008a;105(10):3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F. Lenschow DJ. Zhang DE. Nitrosylation of ISG15 prevents the disulfide bond-mediated dimerization of ISG15 and contributes to effective ISGylation. J Biol Chem. 2008b;283(36):24484–24488. doi: 10.1074/jbc.M803795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F. Zou W. Zhang DE. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 2007;21(3):255–260. doi: 10.1101/gad.1521607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiak A. Utermohlen O. Niendorf S. Horak I. Knobeloch KP. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol Cell Biol. 2005;25(15):6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owhashi M. Taoka Y. Ishii K. Nakazawa S. Uemura H. Kambara H. Identification of a ubiquitin family protein as a novel neutrophil chemotactic factor. Biochem Biophys Res Commun. 2003;309(3):533–539. doi: 10.1016/j.bbrc.2003.08.038. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pincetic A. Kuang Z. Seo EJ. Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol. 2010;84(9):4725–4736. doi: 10.1128/JVI.02478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N. Evans B. Levy D. Fahey D. Knight E., Jr. Darnell JE., Jr Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci USA. 1987;84(18):6394–6398. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KJ. Hahn CS. Kim KI. Yan M. Rosario D. Li L. de la Torre JC. Zhang DE. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10(12):1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- Ritchie KJ. Malakhov MP. Hetherington CJ. Zhou L. Little MT. Malakhova OA. Sipe JC. Orkin SH. Zhang DE. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 2002;16(17):2207–2212. doi: 10.1101/gad.1010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KJ. Zhang DE. ISG15: the immunological kin of ubiquitin. Semin Cell Dev Biol. 2004;15(2):237–246. doi: 10.1016/j.semcdb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS. Dargemont C. Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276(16):12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Satake H. Tamura K. Furihata M. Anchi T. Sakoda H. Kawada C. Iiyama T. Ashida S. Shuin T. The ubiquitin-like molecule interferon-stimulated gene 15 is overexpressed in human prostate cancer. Oncol Rep. 2010;23(1):11–16. [PubMed] [Google Scholar]
- Schwer H. Liu LQ. Zhou L. Little MT. Pan Z. Hetherington CJ. Zhang DE. Cloning and characterization of a novel human ubiquitin-specific protease, a homologue of murine UBP43 (Usp18) Genomics. 2000;65(1):44–52. doi: 10.1006/geno.2000.6148. [DOI] [PubMed] [Google Scholar]
- Shah SJ. Blumen S. Pitha-Rowe I. Kitareewan S. Freemantle SJ. Feng Q. Dmitrovsky E. UBE1L represses PML/RAR{alpha} by targeting the PML domain for ISG15ylation. Mol Cancer Ther. 2008;7(4):905–914. doi: 10.1158/1535-7163.MCT-07-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HX. Yang K. Liu X. Liu XY. Wei B. Shan YF. Zhu LH. Wang C. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol. 2010;30(10):2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T. Iwahara S. Saeki Y. Sasajima H. Yokosawa H. Link between the ubiquitin conjugation system and the ISG15 conjugation system: ISG15 conjugation to the UbcH6 ubiquitin E2 enzyme. J Biochem. 2005;138(6):711–719. doi: 10.1093/jb/mvi172. [DOI] [PubMed] [Google Scholar]
- Takeuchi T. Yokosawa H. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem Biophys Res Commun. 2005;336(1):9–13. doi: 10.1016/j.bbrc.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Tang Y. Zhong G. Zhu L. Liu X. Shan Y. Feng H. Bu Z. Chen H. Wang C. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J Immunol. 2010;184(10):5777–5790. doi: 10.4049/jimmunol.0903588. [DOI] [PubMed] [Google Scholar]
- Tenen DG. Hromas R. Licht JD. Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90(2):489–519. [PubMed] [Google Scholar]
- Versteeg GA. Hale BG. van BS. Wolff T. Lenschow DJ. Garcia-Sastre A. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J Virol. 2010;84(10):5423–5430. doi: 10.1128/JVI.02395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum RR. Ishwaran H. Yoon T. Nuyten DS. Baker SW. Khodarev N. Su AW. Shaikh AY. Roach P. Kreike B. Roizman B. Bergh J. Pawitan Y. van de Vijver MJ. Minn AJ. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105(47):18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Wong JJ. Pung YF. Sze NS. Chin KC. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci USA. 2006;103(28):10735–10740. doi: 10.1073/pnas.0600397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M. Luo JK. Ritchie KJ. Sakai I. Takeuchi K. Ren R. Zhang DE. Ubp43 regulates BCR-ABL leukemogenesis via the type 1 interferon receptor signaling. Blood. 2007;110(1):305–312. doi: 10.1182/blood-2006-07-033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau DA. Hetherington CJ. Wang Q. Zhang P. Sharpe AH. Binder M. Marin-Padilla M. Tenen DG. Speck NA. Zhang DE. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15(3):303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- Yin X. Cong X. Yan M. Zhang DE. Deficiency of a potential 3p21.3 tumor suppressor gene UBE1L (UBA7) does not accelerate lung cancer development in K-rasLA2 mice. Lung Cancer. 2009a;63(2):194–200. doi: 10.1016/j.lungcan.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X. Cong X. Yan M. Zhang DE. Alteration of tumor spectrum by ISGylation in p53-deficient mice. Cancer Biol Ther. 2009b;8(12):1167–1172. doi: 10.4161/cbt.8.12.8558. [DOI] [PubMed] [Google Scholar]
- Yuan W. Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20(3):362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Shin J. Molitor TW. Schook LB. Rutherford MS. Molecular responses of macrophages to porcine reproductive and respiratory syndrome virus infection. Virology. 1999;262(1):152–162. doi: 10.1006/viro.1999.9914. [DOI] [PubMed] [Google Scholar]
- Zhao C. Beaudenon SL. Kelley ML. Waddell MB. Yuan W. Schulman BA. Huibregtse JM. Krug RM. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci USA. 2004;101(20):7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. Denison C. Huibregtse JM. Gygi S. Krug RM. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci USA. 2005;102(29):10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. Hsiang TY. Kuo RL. Krug RM. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc Natl Acad Sci USA. 2010;107(5):2253–2258. doi: 10.1073/pnas.0909144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W. Papov V. Malakhova O. Kim KI. Dao C. Li J. Zhang DE. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem Biophys Res Commun. 2005;336(1):61–68. doi: 10.1016/j.bbrc.2005.08.038. [DOI] [PubMed] [Google Scholar]
- Zou W. Wang J. Zhang DE. Negative regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochem Biophys Res Commun. 2007;354(1):321–327. doi: 10.1016/j.bbrc.2006.12.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W. Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 2006;281(7):3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]