Epigenetic inheritance: Uncontested? (original) (raw)
Abstract
“Epigenetics” is currently defined as “the inheritance of variation (-genetics) above and beyond (epi-) changes in the DNA sequence”. Despite the fact that histones are believed to carry important epigenetic information, little is known about the molecular mechanisms of the inheritance of histone-based epigenetic information, including histone modifications and histone variants. Here we review recent progress and discuss potential models for the mitotic inheritance of histone modifications-based epigenetic information.
Keywords: epigenetics, histone, inheritance, modifications
Introduction
The current definition of “epigenetics” is “the inheritance of variation (-genetics) above and beyond (epi-) changes in the DNA sequence” 1. In eukaryotic cells, the basic unit of chromatin is the nucleosome, composed of 147 bp of DNA wrapped around the core histone octamer 2. Histones contain extensive post-transcriptional modifications for various functions (reviewed in this issue, 3). Histone modifications are known to carry important epigenetic information 3, 4, especially after several histone modifications were shown to be critical to many well-known epigenetic phenomena, including position effect variegation 5, 6, 7, Polycomb silencing 8, 9, 10, 11 and dosage compensation 12, 13. However, how newly deposited histones acquire these modifications during/after DNA replication remains enigmatic. In fact, it is not even clear whether any of these modifications are reestablished on newly deposited histones in a manner precise enough to assure mitotic inheritance of histone modification-based epigenetic information. In this review, we focus on insights obtained from recent progress that lead us to the viewpoint that histone modification-based epigenetic information may not necessarily be precisely inherited during the mitotic divisions. Instead, histone modifications affecting epigenetics may be reestablished in a much more flexible way to maintain distinct transcription states.
Semi-conservative partition, followed by templated modification copying events: an imperfect “ideal” model
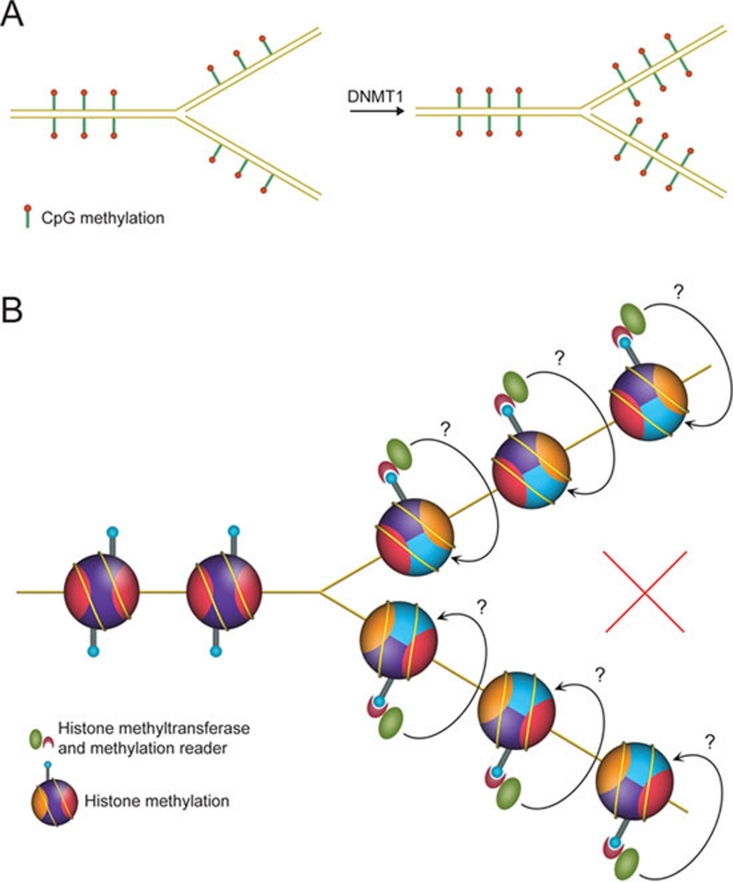
In addition to histone modification-based epigenetic inheritance, there are two well-studied biological phenomena in which information is transmitted across cell division: the inheritance of genetic information through the DNA sequence and the inheritance of epigenetic information encoded by CpG methylation. Semi-conservative DNA replication and sister chromatid segregation ensure the faithful duplication and partition of the genetic information. Similarly, symmetric CpG methylations are segregated in a semi-conservative way and faithfully reestablished on the newly synthesized DNA strand by a templated copying event (Figure 1A) 14, 15, 16. Therefore, semi-conservative partitioning of the histone H3-H4 tetramers followed by templated modification copying events provided a compelling model for the inheritance of histone modifications during mitotic divisions (Figure 1B). This model relies on two critical assumptions: (1) inheritable histone modifications are obligated to exist in a symmetric manner within mono-nucleosomes; (2) histone H3-H4 tetramers undergo semi-conservative segregation during DNA replication-dependent chromatin assembly.
Figure 1.
Semi-conservative partition, followed by templated modification copying events: an imperfect “ideal” model. (A) Model illustration for the inheritance mechanism of DNA methylation. (B) The “ideal” but recently proven incorrect model for the inheritance of histone modifications.
Every nucleosome contains two copies of each core histone. However, it is not known whether histone modifications exist in a symmetric manner within mono-nucleosomes, mainly because of technical difficulties in discriminating symmetrically and asymmetrically modified nucleosomes.
Another key pillar for this precise inheritance model depicted in Figure 1B is the semi-conservative partition of the H3-H4 tetramers during DNA replication-dependent chromatin assembly. Historically, this model about “two symmetrically paired half-nucleosomes” was proposed in 1976 to explain “potential direct inheritance of information in the form of histone structure” 17. However, newly synthesized and parental chromatins exhibit comparable micrococal nuclease digestion kinetics, indicating that newly synthesized chromatin contains full nucleosomes at regular spacing intervals 18. Sedimentation studies with heavy isotope-labeled histones indicated that histone octamers segregate in a conserved manner 19. These findings led to the nucleosome conservative segregation model. Although “hybrid nucleosomes” containing old H3-H4 tetramers and new H2A-H2B dimers, or vice versa, were discovered later 20, 21, 22, 23, H3-H4 tetramers, the core particles of the nucleosomes, which carry the modifications most likely to be involved in epigenetic phenomena, do not dissociate during replication-dependent nucleosome assembly 24, 25, 26, 27. However, expectations for the H3-H4 tetramer split model revived in the past few years, after the discoveries that H3-H4 histones deposit onto chromatin as dimers rather than tetramers 28, 29, 30, and that the histone chaperone Asf1 blocks H3-H4 tetramer formation 31, 32 and even disrupts pre-assembled H3-H4 tetramers to form H3-H4/Asf1 heterotrimers 32. Although without direct evidence, these studies certainly offered a possibility for the H3-H4 tetramer splitting events. Hence, debates and discussions about the H3-H4 tetramer semi-conservative partition model arose in many recent reviews 33, 34, 35, 36, 37, 38, 39. Recently, with selective purification of “old” or “new” H3 histone-containing mono-nucleosomes and stable isotope labeling-based quantitative mass spectrometry analysis, the H3-H4 tetramer partition pattern was reinvestigated in a high-resolution, variant-specific way 40. This study provided strong evidence that the majority of H3-H4 tetramers segregate in a conservative manner. Although splitting events of a fraction of H3.3-H4 tetramers were detected 40, it is unlikely that these events are responsible for epigenetic inheritance mechanisms involving templated modification copying within the same mono-nucleosomes, especially for repressive modifications given that H3.3 is associated with actively transcribed genomic regions.
These observations failed to support the seemingly simple and ideal model for epigenetic inheritance of histone modifications (Figure 1B), leading future investigations into alternative models. These studies also suggest that if there are any templated copying events for histone modifications, the most likely templates would be neighboring pre-modified parental nucleosomes, and, if this were to be the case, then the mitotic inheritance of histone modifications may not be achieved by a highly precise mechanism (see section Templated modification copying events versus modification reinforcement).
Replication-coupled histone modification “copying” versus replication-independent histone modification “maturation”
Replication-coupled modification “copying” on newly deposited histones behind the replication fork?
CpG methylation is faithfully maintained by recruitment of DNMT1 to the replication forks 41 by PCNA 42 and UHRF1 43, 44. Similarly, many histone modifiers have been reported to localize to the replication forks, suggesting potential replication-coupled reestablishment of histone modifications on the nascent chromatin. These include PCNA-mediated replication-coupled recruitment of HDACs 45; CAF-1-mediated MBD1 and SETDB1 recruitment during heterochromatin DNA replication 46, 47, 48; and DNMT1-mediated replication-coupled recruitment of G9a 49. Finally, the PRC2 complex has also been reported to localize to the replication foci throughout S phase 50.
That certain chromatin-modifying enzymes localize to replication foci raises the attractive hypothesis that factors such as PCNA and CAF-1 that interact with the replication machinery may serve as a landing pad for the chromatin-modifying enzymes to facilitate rapid restoration of the histone modifications on nascent chromatin.
Replication-independent modification “maturation” on newly deposited histones?
Histone modifications are often designated as being “epigenetic modifications”. However, certainly not all modifications are “epigenetic”, because many of them may not be inheritable as required under a widely accepted definition of “epigenetics” 1, 4. In fact, even for methylated versions of H3K9 and H3K27 that play critical roles in well-studied epigenetic phenomena, including position effect variegation, Polycomb silencing and X chromosome inactivation in mammals 5, 6, 7, 8, 9, 10, 11, 12, 13, it is not clear whether these modifications are faithfully reestablished on the newly deposited histones. Therefore, the development of a new technology to monitor the establishment of histone modifications on newly deposited histones will help resolve two important questions: (1) which kinds of histone modifications are faithfully reestablished on newly deposited histones; and (2) what is the kinetics for the reestablishment of histone modifications on newly deposited histones?
Stable isotope labeling-based quantitative mass spectrometry analysis allowed simultaneous detection of modifications on pre-existing and newly deposited histones, as well as relative quantification of their global abundance, thus providing a resolution for the above-mentioned technical challenges. This approach was first utilized for studying the reestablishment of H4K20 methylation on newly deposited histones, with the results indicating that H4K20 was progressively methylated throughout the cell cycle 51. Interestingly, little H4K20me2, the main state of mammalian H4K20 methylation, was detected on newly synthesized H4 histones during the S phase, suggesting that H4K20me2 was not reestablished on newly deposited histones in a replication-dependent manner. This was confirmed by an independent study 52. These results are consistent with the findings that H4K20me2-3, established by the Suv4-20 enzymes, requires H4K20me1 53, which is catalyzed by PR-Set7, whose expression is cell cycle-regulated and present from G2 to early G1 54, 55, 56, 57, 58.
A similar strategy was applied to H3K79 methylation, with the improvement that both the H3 histone backbone as well as the methyl group can be simultaneously monitored 59. Surprisingly, old H3 histones continue to be methylated at K79 at a rate comparable to that of newly deposited H3 histones, a clear indication that H3K79 methylation cannot be reestablished in a replication-coupled manner. Given that H3K79me2 shares a similar half-life with the histone modifications most likely to carry epigenetic information (H3K27me3 and H3K9me3) 60, 61, it was proposed that there might be a certain degree of positional “scrambling” of K79 methylation through the cell cycle 59.
Taken together, studies with stable isotope labeling-based quantitative mass spectrometry analysis provided new technology and novel insights for understanding the modification-reestablishment process on newly deposited histones. Current results indicated that histone lysine methylations in general have a much longer half-life than acetylation or phosphorylation 51, 60, 61, and that certain methylation states might be established on newly deposited histones in a chromatin assembly-coupled way. But clearly, modification states like H4K20me2 are established by a mechanism independent of DNA replication 51, 52. These modification states might gradually “mature” through the cell cycle.
Templated modification copying events versus modification reinforcement
Templated modification copying events?
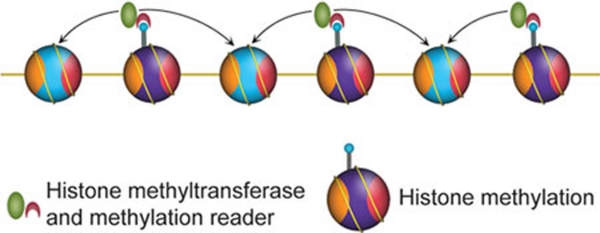
In order to be heritable, pre-existing modifications should, at least to some extent, be able to serve as templates to guide the reestablishment of the same modifications on newly deposited histones 62. Considering that the vast majority of histone H3-H4 tetramers segregate in a conservative manner 26, 27, 40, the most likely templates might be adjacent pre-existing nucleosomes in cis (Figure 2).
Figure 2.
Modification spreading model that can also be involved in the epigenetic inheritance of histone modifications.
The proposed modification copying events are highly similar to another analogous phenomenon, chromatin modification spreading. Certain histone modification events, including H4K16 deacetylation, H3K9 methylation and H3K27 methylation, can spread along the chromatin to maintain a regional heterochromatic environment 63, 64, 65. These events are typically mediated by the association of a chromatin-modifying enzyme with a partner effector protein that specifically recognizes the modified product, such as Sir2 (H4K16 deacetylase) 66 with Sir3 (H4K16 effector) 67, Suvar39/Clr4 (H3K9 methyltransferase) 5, 68 with HP1/Swi6 (H3K9me3 effector) 6, 7, 68, and PRC2-Ezh2 (H3K27 methyltransferase) 8, 9, 10, 11 with Eed, a subunit of PRC2 as the H3K27me3 effector 50, 65. Therefore, inheritance of histone modifications can be achieved through similar biochemical mechanisms.
A recent study describing allosteric activation of PRC2 by H3K27me3 peptide suggests that PRC2 can “sense” the neighboring repressive environment and intrinsically mediate chromatin modification propagation 65. A similar strategy might be used for other histone-modifying enzymes, although stimulation of histone methyltransferase activities by neighboring methylated nucleosomes in cis awaits biochemical demonstration. Importantly, this mechanism is compatible with both the replication-coupled modification “copying” model and the replication-independent modification “maturation” model.
Modification reinforcement?
Although the above-mentioned templated modification copying events appear practical, at least for some modifications, accumulating evidence suggests many epigenetic states may not be inherited solely by this simple mechanism.
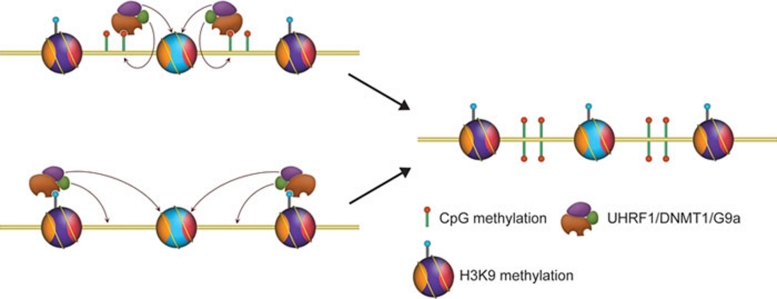
Several chromatin modifications may act coordinately to maintain the epigenetic states reflected by these modifications. One good example is the trio among UHRF1, DNMT1 and H3K9 methylation (Figure 3). UHRF1 recruits DNMT1 to hemimethylated CpG sites during DNA replication to facilitate the maintenance of DNA methylation 43, 44; DNMT1 interacts with G9a and recruits it to replication foci, potentially for replication-coupled H3K9me2 reestablishment on the nascent chromatin 49. Interestingly, H3K9me2 catalyzed by G9a or H3K9me3 catalyzed by Su(var)39-h can also recruit UHRF1 through direct recognition 69 and potentially facilitate DNA methylation. This is a clear feedback loop with players in this system reinforcing each other for eventual maintenance of the epigenetic states that they present. We expect that similar cases for other chromatin modifications will be discovered in the coming years.
Figure 3.
A reinforcement model for epigenetic inheritance. See text for details.
Finally, we would like to point out that the transcription status of a gene locus itself might play a role in maintaining its epigenetic status. Numerous chromatin modifiers are known to associate with the transcription machinery (for a review, see 70); therefore, it is easily conceivable that active transcription maintained by these modifications may in turn facilitate the reestablishment of these modifications in the next cell cycle. Although less apparent, it is also likely that transcription repression mediated by repressive modifications may also in turn facilitate the reestablishment of these modifications by certain favorable characteristics of the repressed chromatin, which awaits further investigations.
Concluding remarks
In comparison to genetic information which is strictly inherited via “code-copying” (DNA replication), recent progress suggests that epigenetic inheritance mechanisms are much more complicated. Although the mechanisms underlying epigenetic inheritance appear to be far less faithful or rigid in comparison to DNA replication, they do in fact offer an opportunity for plasticity, which is critical to the most fundamental role of epigenetic information, which is to translate one genome into hundreds of epigenomes for the generation of distinct cellular functions.
Acknowledgments
Work in the laboratory of BZ is supported by grants from the Chinese Ministry of Science and Technology (863 project 2007AA02Z1A6 and 973 project 2011CB965300); work in the laboratory of DR is funded by NIH (grants RO1GM064844 and 4R37GM037120) and the Howard Hughes Medical Institute.
References
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Thomas JO. Chromatin structure: oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T.Regulation of chromatin by histone modifications Cell Res 2011. Feb 15. doi: 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed]
- Allis CD, Jenuwein T, Reinberg D.Overviews and conceptsIn: Allis CD, Jenuwein T, Reinberg D, eds. Epigenetics. New York: Cold Spring Harbor Laboratory Press, 200623–56.
- Rea S, Eisenhaber F, O'Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Worcel A, Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976;9:409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- Seale RL. Studies on the mode of segregation of histone nu bodies during replication in HeLa cells. Cell. 1976;9:423–429. doi: 10.1016/0092-8674(76)90087-8. [DOI] [PubMed] [Google Scholar]
- Leffak IM, Grainger R, Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977;12:837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Russev G, Hancock R. Formation of hybrid nucleosomes containing new and old histones. Nucleic Acids Res. 1981;9:4129–4137. doi: 10.1093/nar/9.16.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981;23:121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. A reevaluation of new histone deposition on replicating chromatin. J Biol Chem. 1981;256:5095–5103. [PubMed] [Google Scholar]
- Annunziato AT, Schindler RK, Riggs MG, Seale RL. Association of newly synthesized histones with replicating and nonreplicating regions of chromatin. J Biol Chem. 1982;257:8507–8515. [PubMed] [Google Scholar]
- Prior CP, Canto CR, Johnson EM, Allfrey VG. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980;20:597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- Jackson V. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry. 1988;27:2109–2120. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990;29:719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Yamasu K, Senshu T. Conservative segregation of tetrameric units of H3 and H4 histones during nucleosome replication. J Biochem. 1990;107:15–20. doi: 10.1093/oxfordjournals.jbchem.a122999. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- English CM, Maluf NK, Tripet B, Churchill ME, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson LJ, Gu Y, Yakovleva T, et al. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperon CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Annunziato AT. Split decision: what happens to nucleosomes during DNA replication. J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Ray-Gallet D, Quivy JP, Tagami H, Almouzni G. Two distinct nucleosome assembly pathways: dependent or independent of DNA synthesis promoted by histone H3.1 and H3.3 complexes. Cold Spring Harb Symp Quant Biol. 2004;69:273–280. doi: 10.1101/sqb.2004.69.273. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc Natl Acad Sci USA. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Milutinovic S, Zhuang Q, Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J Biol Chem. 2002;277:20974–20978. doi: 10.1074/jbc.M202504200. [DOI] [PubMed] [Google Scholar]
- Reese BE, Bachman KE, Baylin SB, Rountree MR. The methyl-CpG binding protein MBD1 interacts with the p150 subunit of chromatin assembly factor 1. Mol Cell Biol. 2003;23:3226–3236. doi: 10.1128/MCB.23.9.3226-3236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Loyola A, Tagami H, Bonaldi T, et al. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estève PO, Chin HG, Smallwood A, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf AN, Barth TK, Imhof A. Establishment of histone modifications after chromatin assembly. Nucleic Acids Res. 2009;37:5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hübner MR, Beck DB, et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4 Cdt2-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wang W, Kong X, et al. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010;24:2531–2542. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, et al. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell. 2010;40:22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- Sweet SM, Li M, Thomas PM, Durbin KR, Kelleher NL. Kinetics of re-establishing H3 K79 methylation marks in global human chromatin. J Biol Chem. 2010;285:32778–32786. doi: 10.1074/jbc.M110.145094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Xu M, Zhu B. Nucleosome assembly and epigenetic inheritance. Protein Cell. 2010;1:820–829. doi: 10.1007/s13238-010-0104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M, Gotta M, Palladino F, Martin SG, Gasser SM. Targeting Sir proteins to sites of action: a general mechanism for regulated repression. Cold Spring Harb Symp Quant Biol. 1998;63:401–412. doi: 10.1101/sqb.1998.63.401. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]