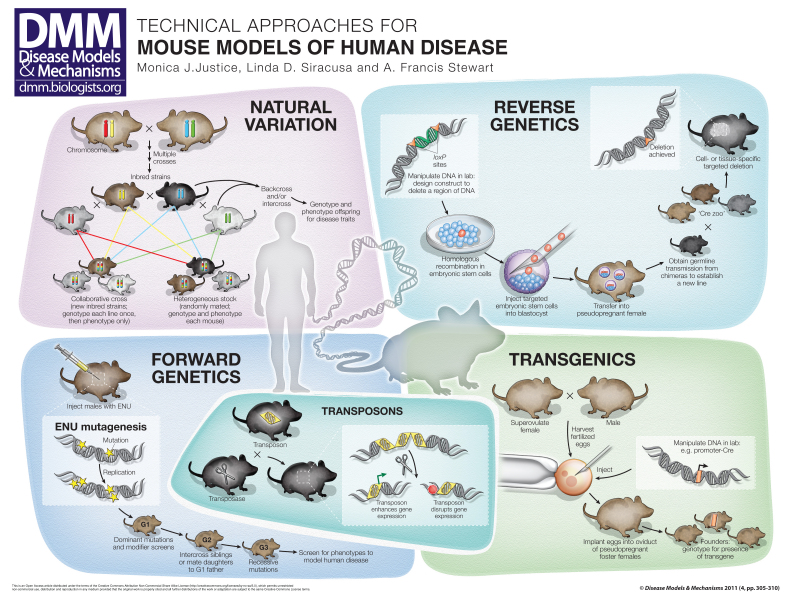
Technical approaches for mouse models of human disease (original) (raw)
Abstract
The mouse is the leading organism for disease research. A rich resource of genetic variation occurs naturally in inbred and special strains owing to spontaneous mutations. However, one can also obtain desired gene mutations by using the following processes: targeted mutations that eliminate function in the whole organism or in a specific tissue; forward genetic screens using chemicals or transposons; or the introduction of exogenous transgenes as DNAs, bacterial artificial chromosomes (BACs) or reporter constructs. The mouse is the only mammal that provides such a rich resource of genetic diversity coupled with the potential for extensive genome manipulation, and is therefore a powerful application for modeling human disease. This poster review outlines the major genome manipulations available in the mouse that are used to understand human disease: natural variation, reverse genetics, forward genetics, transgenics and transposons. Each of these applications will be essential for understanding the diversity that is being discovered within the human population.
Natural variation
Natural variation is a change in one or more nucleotides between individuals. The change can be a simple substitution or a complex rearrangement. Deletions, point mutations, amplifications, insertions, inversions and translocations can alter the DNA sequence. Such changes occur by chance and hence are classified as spontaneous mutations. Transmission from parent to offspring results in fixation of these mutations within a population.
As schematized in the poster, mice caught in the wild can contain one of four bases (A, C, G or T) at a given locus; a female and male are initially mated in the laboratory to produce offspring. Inbreeding by brother-sister matings for 20 consecutive generations creates inbred strains (see Box 1 for definition of terms) of mice, and each individual strain is virtually homozygous for every gene in the genome (isogenic) (Silver, 1995). Inbred strains have thus captured a portion of the natural variation present in wild mice. The Mouse Genome Database describes the origins and characteristics of inbred strains and is a comprehensive database for information about the mouse (www.informatics.jax.org). Given the large number of inbred strains, it is estimated that these collective resources now exhibit more variation than that present in the human population.
Box 1.
Glossary
Alleles:
an array of possible forms of a gene, which can each cause different phenotypic effects.
Backcross:
the mating of a heterozygous individual with one of its inbred parents, or with an individual of the same genotype as one of its inbred parents, to follow the inheritance of alleles and phenotypes.
Congenic strain:
a strain derived by backcrossing an allele or mutation of interest onto the background of a different strain for at least ten generations to achieve allelic transfer.
Conplastic strain:
a strain that carries the mitochondrial genome from a donor strain and the nuclear genome of a recipient strain. Conplastic strains are made by intercrossing two inbred strains, followed by sequential crosses of females to recipient males.
Consomic strain:
a strain that carries one pair of homologous whole chromosomes from a donor strain on the genetic background of a recipient strain. A complete set of consomic strains is 21, which includes those with each of the 19 autosome pairs, the X chromosome pair and the Y chromosome.
Forward genetics:
a genetic analysis that proceeds from phenotype to genotype by positional cloning or candidate gene analysis.
Inbred strain:
a strain derived by at least 20 generations of sequential brother by sister matings.
Intercross:
a cross between two individuals with the same heterozygous genotype (usually a brother by sister mating), to follow the inheritance of alleles and phenotypes.
Recombinant congenic (RC) strains:
a set of strains derived from intercrossing two inbred strains, followed by a small number of backcross generations prior to inbreeding. A subset of the donor genome remains present on the background of the recipient strain.
Recombinant inbred (RI) strains:
a set of strains derived from intercrossing two inbred strains, then brother by sister mating their F2 offspring for at least 20 generations to derive new inbred strains. Each strain represents a random mixture of genes from the two parental strains that are fixed in the new inbred strain.
Reverse genetics:
A genetic analysis that proceeds from genotype to phenotype by genetic engineering techniques, such as homologous recombination in embryonic stem (ES) cells.
Natural variation accounts for the differences in phenotype exhibited among inbred strains (as well as between individuals). Sequence differences can alter transcripts and proteins by altering their functional properties, as well as the timing, level and site(s) of expression in the body. Monogenic traits are controlled by only one gene, whereas complex traits are controlled by two or more genes. Intercrosses and backcrosses between inbred strains exhibiting the extremes of a specific phenotype permit a general localization of genes influencing the trait by matching phenotype to genotype. Such crosses have also been used to identify ‘modifier’ genes, namely genes that can modify the phenotype of an already existing mutation or disease allele; thus, a disease can manifest as mild or severe, depending on the influence of modifier alleles (Nadeau, 2003). The chromosomal location of each type of gene can be refined through the use of consomic, conplastic, congenic, recombinant inbred (RI) and/or recombinant congenic (RC) strains (Roberts et al., 2007).
An electronic version of this poster is available online.
Natural variation can be used to identify genes involved in a variety of quantitative traits and diseases (Hunter and Crawford, 2008). Already established and developing resources are designed to mimic the diversity present in human populations. The Collaborative Cross (CC) is a combination of the genotypes of eight inbred strains, specifically chosen because of the diversity of their genomes (only four strains are shown in the poster for simplicity) (Churchill et al., 2004). More than 500 individual inbred strains derived from the CC will be completed soon. Each of the eight inbred strains that contribute to the CC is being sequenced, so that the molecular genetic contribution of each chromosomal region will be known by determining the genotype of each CC strain using the mouse diversity chip (Yang et al., 2009). Because the molecular composition of each CC strain will be known, phenotyping each strain for a trait such as immune response, blood glucose level, bone density or social behavior will allow researchers to determine a refined chromosomal location for quantitative trait loci (QTLs) affecting the trait of interest using in silico methods. An advantage of the CC is that all data are cumulative, allowing for cross-referencing of phenotype and genotype databases derived from the CC strains.
Heterogeneous stock (HS) populations also represent contributions from eight inbred strains; however, HS populations are maintained by random matings of mice within a colony (Valdar et al., 2006). Therefore, although the molecular contribution from the eight progenitor inbred strains is known, new inbred strains are not created. Thus, each individual HS mouse has a unique combination of alleles (see poster). HS mice are designed to contain random variation within a population that would be similar to that found in the human population, allowing for a controlled molecular knowledge of the alleles that contribute to each mouse. Because each individual derived from the HS cross is unique, the molecular component of each mouse must be genotyped, and each mouse must be phenotyped to identify genes affecting the trait of interest. The benefit of these novel resources is the ability to map a trait of interest to a small chromosomal region, and ultimately to identify the genes and pathways responsible so as to better prevent and treat human disease.
Current sequencing efforts for the human genome are revealing a large number of genetic alterations that occur from generation to generation. The nature of these ‘private’ mutations and their impact on phenotype has yet to be fully evaluated. Such individual variation contributes to a phenotype by uniquely affecting the individual who carries such changes. These types of changes can be evaluated in the mouse by using reverse genetics or transgenesis.
Reverse genetics
The ability to introduce a mutation by design is termed reverse genetics. Of all experimental organisms, fluent reverse genetics is currently only available in Escherichia coli, yeast and the mouse, owing to the remarkable renewing properties of mouse embryonic stem cells (mESCs) (Soriano, 1995). This feature is one of the main reasons for the pre-eminence of the mouse as the leading mammalian model system (Oliver et al., 2007). mESCs are derived from early-stage embryos and can be grown in culture yet retain the ability to contribute to the formation of a mouse on reintroduction into an early-stage host embryo – usually at the blastocyst stage, as illustrated in the poster. Because mESCs can be grown in culture, the mouse genome can be engineered by transfection with DNA constructs that have been engineered in vitro to carry a specifically designed mutation (the insertion of loxP sites is shown in the poster). The DNA construct can be precisely integrated into the mouse genome via homologous recombination, so that the existing DNA sequence is altered exactly by design (Capecchi, 1989). Such constructs are commonly designed to ‘knock out’ the gene by deleting or replacing coding exon(s). Alternatively, the gene can be disrupted by random or transpositional ‘gene-trapping’ mutagenesis (Evans et al., 1997). Gene trapping varies from targeted mutagenesis in that a construct carrying a reporter gene can insert into a gene, disrupting its function. The sequence flanking the insertion is determined subsequently, and the expression of the reporter can allow for the assessment of where the gene is expressed. The advantage of gene trapping is the speed at which genes can be disrupted. A disadvantage is that the insertions might not disrupt the gene, or that they have the potential to affect flanking genes owing to their position.
For either targeted mutations or gene-trapped mutations, transfected mESC clones can be selected after screening for disrupted alleles in the gene of interest. After injection into a blastocyst, the selected mESC clone mixes into the host embryo to contribute to a chimera composed of host and mutated cells; this chimeric adult can propagate the mESC mutation by germline transmission to create a genetically engineered mouse strain.
The ability to engineer the mouse genome via mESCs stimulated the development of sophisticated genome engineering and gene expression strategies (Glaser et al., 2005). Chief among these is conditional mutagenesis based on the use of Cre-loxP site-specific recombination (Branda and Dymecki, 2004). Two 34-bp loxP sites are introduced into a chosen gene, either by gene targeting or trapping, so that normal gene expression is undisturbed. The ‘floxed’ gene is mutagenized upon exposure to Cre recombinase, which mediates DNA recombination between the loxP sites. The mutagenic event can be controlled in space and time by regulating Cre recombinase expression and/or activity (Schnutgen et al., 2006). For example, it could be that complete elimination of a gene of interest results in embryonic lethality due to a requirement in embryogenesis; however, mating a conditional allele of this gene to a Cre expresser during adulthood would result in the elimination of the gene long after it is required for embryonic development, allowing the researcher to investigate the function of the gene in the adult.
A growing panel of mouse lines that express Cre recombinase in specific cell types, termed the ‘Cre zoo’, is being developed to achieve spatial precision in mutagenesis (see Transgenics section). Additionally, temporal regulation of Cre recombinase activity can be achieved by expressing Cre as a fusion protein with a steroid receptor ligand-binding domain, usually the mutant estrogen domain termed ‘ERT2’, which is activated by 4-hydroxytamoxifen. Thus, crossing a Cre driver mouse with a mouse carrying a floxed gene enables conditional mutagenesis in an inducible way. Some technical considerations for targeted mutagenesis are outlined in Box 2.
Box 2. Technical considerations for targeted mutagenesis.
- To select for the construct in mESCs, selection cassettes must be incorporated. For example, neomycin allows cells to grow in media that contains G418, and hypoxanthine phosphoribosyltransferase (HPRT) allows growth in medium containing hypoxanthine, aminopterin and thymidine (HAT). Growing the cells in such media is required, but repeated selection can be detrimental to the cells (Plagge et al., 2000). Furthermore, the presence of internal promoters required to drive the expression of selection cassettes can influence the expression of flanking genes. Thus, removal of the cassettes is desired prior to the analysis of phenotype. This can be achieved by the use of another site-specific recombination system derived from yeast: the Flp recombinase catalyzes recombination at frt sites (Farley et al., 2000).
- The construct will undergo homologous recombination more readily in an isogenic background. The most commonly used mESCs are AB2.2, which are derived from the 129/Ola inbred strain, and JM8, which are derived from the C57BL/6N inbred strain. The availability of knockouts on either strain background reduces the likelihood that modifier genes will affect the phenotype after mice are made. However, each strain background has different germline transmission rates (Pettitt et al., 2009).
- Germline transmission of the mESC genetic component is required to obtain mice that carry the construct. However, aneuploidy of the mESCs can preclude the ability of the clone to achieve germline transmission. Obtaining early passage mESCs and reducing the manipulations required to generate the desired construct can make germline transmission more likely.
- Human disease can also be caused by lesions other than deletions that alter gene function. For example, point mutations can be modeled using a ‘knock-in’, whereby the precise lesion found in the human is introduced into the mouse using a two-step procedure in mESCs (Plagge et al., 2000). Alternatively, a transgene expressing the human disease mutation can be introduced into the knockout background.
Transgenics
A gene that does not inherently belong to the organism, but is introduced into its genome from an outside source is called a transgene. Transgenesis can be achieved in mice by the microinjection of manipulated DNA into the pronucleus of a fertilized egg (Palmiter and Brinster, 1985). The DNA inserts at random into the genome, and if it does so prior to cell division, it can contribute to all cells of the organism. Exogenous transposons as well as engineered or targeted mutations are also transgenes; however, these are not examples of microinjection transgenics.
To obtain ova for microinjection, female mice must be superovulated using hormone injections to obtain a maximum number of embryos after mating to males. Fertilized eggs are harvested and injected with linearized DNA constructs in the laboratory, then surgically implanted into the oviducts of foster mothers that are ‘pseudopregnant’, having been mated with vasectomized males to induce appropriate hormones for pregnancy. The subsequent liveborn mice represent ‘founders’ carrying the transgene; these mice must be analyzed for the presence or absence of the exogenous DNA.
The first application of mouse transgenesis was a physiological one: the introduction of human growth hormone resulted in increased size (Palmiter et al., 1982). Subsequent applications of transgenesis were to express a gene under the control of exogenous regulatory elements, or to drive a reporter gene under the control of regulatory elements. Such experiments have been very powerful for cancer studies, as well as for understanding mechanisms of gene regulation.
Shown in the poster is an example of a Cre driver mouse: a promoter element is used to drive expression of Cre recombinase in a spatial- or temporal-specific pattern. As explained earlier, in the reverse genetics section, Cre drivers are required for conditional deletion of a region of DNA from mice that contain loxP sites. Other applications of transgenics are to express a bacterial artificial chromosome (BAC) or yeast artificial chromosome (YAC) in the mouse, for gene rescue or for ‘humanization’ (expression of a human gene or genomic region in the mouse).
There are several problems with microinjection transgenics, which must be understood for their proper use.
- Because integration is random, endogenous regulatory elements can influence the expression of the transgene.
- The linearized DNA commonly integrates as a head-to-tail concatamer in copy numbers that vary from one to >200 per haploid genome. Copy number influences the level of expression, and copies can be lost in subsequent generations by gene conversion.
- If the DNA integrates late in embryonic cell division, the founder mice can be mosaic, and must be bred to achieve contribution of the transgene to all cells. This is a common problem for large transgenes such as BACs.
- Integration can result in an insertional mutation, as demonstrated in the transposon section (see below).
Each of these issues can influence expression or phenotypic outcome. Thus, making microinjection transgenic mice requires the analysis of multiple founder lines. Recent techniques using site-specific recombinases obtained from bacteriophages have been developed to alleviate these concerns. These include the use of ‘docking sites’ for site-specific integrases so that the effect of position and copy number is reduced, thus eliminating the requirement for multiple founders (Branda and Dymecki, 2004).
An adaptation of Cre-loxP conditional expression uses knock-in alleles to overcome many of the disadvantages of classical transgenics. A gene can be placed under the control of a ubiquitous promoter (driving the expression of the gene in all cells at all times) or self promoter (driving the expression of the gene under the control of its normal promoter), but transcriptional STOP sequences, which are flanked by loxP sites, can be inserted to block its expression. When the animal is mated to a Cre expresser, or Cre is spatially introduced (as in viral Cre delivery), recombination occurs between the loxP sites, allowing for deletion of the STOP cassette, inducing expression of the gene in a tissue or site of interest (Zadelaar et al., 2006). This application is powerful for cancer studies, because expression of a single copy gene is precisely regulated by time and location; it can be designed to allow expression in a single cell (Marumoto et al., 2009).
Forward genetics
In forward genetics approaches, many agents, including chemicals, radiation and viruses, are used to disrupt genes to identify their functions and the diseases associated with them. The most powerful mutagen for forward genetic screens in mice is the alkylating agent _N_-ethyl-_N_-nitrosourea (ENU), which can produce mutation rates as high as 1.5×10–3 per mutagenized genome in male mouse spermatogonial stem cells (Russell et al., 1979; Guenet, 2005). ENU primarily produces point mutations, which include loss-of-function, gain-of-function, and super-active and partially active coding region mutations, as well as non-coding RNA and regulatory mutations.
After treatment with ENU, male mice are mated in genetic screens designed to uncover mutations of interest (Justice, 2000). Dominant mutations are isolated by their phenotype in the first generation of breeding (Hrabe de Angelis and Balling, 1998). Mutations that result in visible phenotypes, such as changes in the coat, morphology or movement, are simple to detect. More advanced phenotype screening methods for behavior, hematology, pain perception and biochemistry have uncovered many previously unknown dominant mutations. Screens for modifying mutations are likely to be the most common use of forward genetics in the future, because they are a powerful method for identifying disease suppressors (Carpinelli et al., 2004). In a modifier screen, a new unknown dominant mutation present in the ENU-treated male gamete is isolated by its ability to modify (either by enhancing or suppressing) a known recessive or dominant phenotype that is produced by a mutation carried by a female mouse with which the male is mated, or that is present as a homozygous viable trait in the strain background. Here, the idea of complex traits is taken to the extreme: instead of relying on natural variation, potent mutations are induced in DNA, and the animal reveals important interactions.
Screens for recessive mutations using three-generation pedigree breeding schemes or using balancer chromosomes have been carried out (Kasarskis et al., 1998; Herron et al., 2002; Kile et al., 2003). Although recessive screens require more breeding time, their use has produced mutations to understand developmental pathways, immunology and responses to infection (Beutler et al., 2007; Stottmann and Beier, 2010). In all cases, a mutation is mapped to a molecular interval, and then genes are sequenced to identify lesions caused by ENU treatment and not repaired.
Banks of sperm and DNA samples from mutagenized males are useful for identifying point mutations in desired genes (Coghill et al., 2002). The identification of point mutations is now straightforward owing to advances in sequencing technology and in mutation detection. Once a mutation is identified, the affected gene must be confirmed by a second mutation, perhaps by a gene knockout, or by a rescue of the mutant phenotype using a transgene.
Transposons
Transposable elements are discrete pieces of DNA that can ‘jump around’ in the genome of a living organism. For each DNA transposon, a corresponding protein, called a transposase, mediates the jumping. Two exogenous transposon-transposase duos have proven to be powerful forward genetic mutagens in the mouse: Sleeping Beauty (SB), which was a non-functioning system that has been resurrected from salmonid fish, and PiggyBac, which is native to the cabbage looper moth (Bestor, 2005; Horie et al., 2010). Unlike ENU mutagenesis, which requires large amounts of sequencing to pinpoint each tiny change, transposable elements are powerful because their sequence is known, so when a transposon insertion mutates a gene, the transposon sequence provides a ‘tag’ to quickly pinpoint its location in the vast sea of genomic DNA. Each of these elements works by a ‘cut-and-paste’ mechanism so that the transposase (represented by scissors in the poster) cuts the transposon out of one location, allowing the transposon to hop to a new location. The transposon is flanked by direct repeats (represented by arrowheads), which are required for transposition. In its new location, the transposon can enhance gene expression (green arrow), disrupt gene expression (stop sign), or have no effect on gene expression. The most powerful application of the transposon systems in mice has been to identify genes that promote cancer (Collier and Largaespada, 2007). The system has applications outside mutagenesis, because the transposons can be engineered to deliver DNA cargo to many locations in the genome using the direct repeat sequences. Thus, the system overlaps transgenic and forward-genetic methods.
Moving forward
Genome-wide association studies (GWAS) are continually identifying a growing number of loci that are associated with human diseases [see the catalog compiled by the National Human Genome Research Institute (NHGRI) at www.genome.gov/gwastudies/index.cfm], increasing the need to investigate the function of these disease alleles in vivo. Furthermore, whole-genome sequencing is just beginning to reveal what might be a multitude of rare, individual mutations that underlie human disease and other complex traits (Manolio et al., 2009). The only means to understand biological processes that occur in cells and physiological processes that occur in whole organisms is to recapitulate alleles associated with disease in a controlled setting, and to explore the effects of selected mutations and/or variants, both individually and in combination. The mouse is the only mammalian system that currently has the resources as well as the technology to fulfill this challenge. The combination of forward and reverse genetics, along with transgenesis and the ability to exploit natural variation, provides a variety of approaches to selectively manipulate the mouse genome and reveal the impact of genetic variants as well as discover missing heritability factors. To move the field forwards on a large scale, the International Knockout Mouse Consortium (IKMC) will soon achieve its goal of having a mouse mutant or a targeted mESC for every gene, providing a crucial resource for functional annotation (www.knockoutmouse.org). Mouse disease clinics, which use broad-based phenotyping platforms, will assess these strains to understand gene function and model human disease (http://eumodic.org; http://nihroadmap.nih.gov/KOMP2/). The Complex Trait Community will soon generate up to 1000 strains that can be phenotyped for many traits and under many conditions (http://www.complextrait.org/). In support of these efforts, 45 of the most commonly used inbred strains that show a high degree of diversity are being analyzed for common variation or are being sequenced, providing powerful information on the origin and evolution of inbred strains of mice (http://www.ensembl.org/Mus_musculus/Info/Index).
The challenge now is to expand our creativity by developing novel assays to detect a broad spectrum of phenotypes that are relevant to human health and disease. Studies in mice and humans are thus poised to inform the other, resulting in new avenues for disease prevention, risk assessment, diagnosis and treatment. The future holds great promise as we follow the unwinding DNA road towards a fuller understanding of the genetic basis for human health.
REFERENCES
- Bestor T. H. (2005). Transposons reanimated in mice. Cell 122, 322–325 [DOI] [PubMed] [Google Scholar]
- Beutler B., Du X., Xia Y. (2007). Precis on forward genetics in mice. Nat. Immunol. 8, 659–664 [DOI] [PubMed] [Google Scholar]
- Branda C. S., Dymecki S. M. (2004). Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6, 7–28 [DOI] [PubMed] [Google Scholar]
- Capecchi M. R. (1989). The new mouse genetics: altering the genome by gene targeting. Trends Genet. 5, 70–76 [DOI] [PubMed] [Google Scholar]
- Carpinelli M. R., Hilton D. J., Metcalf D., Antonchuk J. L., Hyland C. D., Mifsud S. L., Di Rago L., Hilton A. A., Willson T. A., Roberts A. W., et al. (2004). Suppressor screen in Mpl-/- mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl. Acad. Sci. USA 101, 6553–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., Beatty J., Beavis W. D., Belknap J. K., Bennett B., Berrettini W., et al. (2004). The collaborative cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36, 1133–1137 [DOI] [PubMed] [Google Scholar]
- Coghill E. L., Hugill A., Parkinson A., Davison C., Glenister P., Clements S., Hunter J., Cox R. D., Brown S. D. (2002). A gene-driven approach to the identification of ENU mutants in the mouse. Nat. Genet. 30, 255–256 [DOI] [PubMed] [Google Scholar]
- Collier L. S., Largaespada D. A. (2007). Transposons for cancer gene discovery: Sleeping Beauty and beyond. Genome Biol. 8 Suppl. 1, S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J., Carlton M. B. L., Russ A. P. (1997). Gene trapping and functional genomics. Trends Genet. 13, 370–374 [DOI] [PubMed] [Google Scholar]
- Farley F. W., Soriano P., Steffen L. S., Dymecki S. M. (2000). Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110 [PubMed] [Google Scholar]
- Glaser S., Anastassiadis K., Stewart A. F. (2005). Current issues in mouse genome engineering. Nat. Genet. 37, 1187–1193 [DOI] [PubMed] [Google Scholar]
- Guenet J.-L. (2005). Chemical mutagenesis of the mouse genome: an overview. In Genetica: Mutagenesis of the Mouse Genome (ed. Justice M. J., Bedell M. A.), pp. 9–24 Dordrecht, The Netherlands: Klewer Academic Publications; [PubMed] [Google Scholar]
- Herron B. J., Lu W., Rao C., Liu S., Peters H., Bronson R. T., Justice M. J., McDonald J. D., Beier D. R. (2002). Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat. Genet. 30, 185–189 [DOI] [PubMed] [Google Scholar]
- Horie K., Kokubu C., Takeda J. (2010). Functional genomics in the mouse using the sleeping beauty transposon system. Methods Enzymol. 477, 71–89 [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M., Balling R. (1998). Large scale ENU screens in the mouse: genetics meets genomics. Mutat. Res. 400, 25–32 [DOI] [PubMed] [Google Scholar]
- Hunter K. W., Crawford N. P. (2008). The future of mouse QTL mapping to diagnose disease in mice in the age of whole-genome association studies. Annu. Rev. Genet. 42, 131–141 [DOI] [PubMed] [Google Scholar]
- Justice M. J. (2000). Capitalizing on large-scale mouse mutagenesis screens. Nat. Rev. Genet. 1, 109–115 [DOI] [PubMed] [Google Scholar]
- Kasarskis A., Manova K., Anderson K. V. (1998). A phenotype-based screen for embryonic lethal mutations in the mouse. Proc. Nat. Acad. Sci. USA 95, 7485–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile B. T., Hentges K. E., Clark A. T., Nakamura H., Salinger A. P., Liu B., Box N., Stockton D. W., Johnson R. L., Behringer R. R., et al. (2003). Functional genetic analysis of mouse chromosome 11. Nature 425, 81–86 [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., Hunter D. J., McCarthy M. I., Ramos E. M., Cardon L. R., Chakravarti A., et al. (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T., Tashiro A., Friedmann-Morvinski D., Scadeng M., Soda Y., Gage F. H., Verma I. M. (2009). Development of a novel mouse glioma model using lentiviral vectors. Nat. Med. 15, 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. H. (2003). Modifier genes and protective alleles in humans and mice. Curr. Opin. Genet. Dev. 13, 290–295 [DOI] [PubMed] [Google Scholar]
- Oliver P. L., Bitoun E., Davies K. E. (2007). Comparative genetic analysis: the utility of mouse genetic systems for studying human monogenic disease. Mamm. Genome 18, 412–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D., Brinster R. L. (1985). Transgenic mice. Cell 41, 343–345 [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Brinster R. L., Hammer R. E., Trumbauer M. E., Rosenfeld M. G., Birnberg N. C., Evans R. M. (1982). Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature 300, 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt S. J., Liang Q., Rairdan X. Y., Moran J. L., Prosser H. M., Beier D. R., Lloyd K. C., Bradley A., Skarnes W. C. (2009). Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods 6, 493–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagge A., Kelsey G., Allen N. D. (2000). Directed mutagenesis in embryonic stem cells. In Mouse Genetics and Transgenics: A Practical Approach (ed. Jackson I. J., Abbott C. M.), pp. 247–284 Oxford, Oxford University Press [Google Scholar]
- Roberts A., Pardo-Manuel de Villena F., Wang W., McMillan L., Threadgill D. W. (2007). The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm. Genome 18, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W. L., Kelly E. M., Hunsicker P. R., Bangham J. W., Maddux S. C., Phipps E. L. (1979). Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl. Acad. Sci. USA 76, 5818–5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnutgen F., Stewart A. F., von Melchner H., Anastassiadis K. (2006). Engineering embryonic stem cells with recombinase systems. Methods Enzymol. 420, 100–136 [DOI] [PubMed] [Google Scholar]
- Silver L. M. (1995). In Mouse Genetics, Concepts and Applications. New York: Oxford University Press [Google Scholar]
- Soriano P. (1995). Gene targeting in ES cells. Annu. Rev. Neurosci. 18, 1–18 [DOI] [PubMed] [Google Scholar]
- Stottmann R. W., Beier D. R. (2010). Using ENU mutagenesis for phenotype-driven analysis of the mouse. Methods Enzymol. 477, 329–348 [DOI] [PubMed] [Google Scholar]
- Valdar W., Solberg L. C., Gauguier D., Burnett S., Klenerman P., Cookson W. O., Taylor M. S., Rawlins J. N. P., Mott R., Flint J. (2006). Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38, 879–887 [DOI] [PubMed] [Google Scholar]
- Yang H., Ding Y., Hutchins L. N., Szatkiewicz J., Bell T. A., Paigen B. J., Graber J. H., de Villena F. P., Churchill G. A. (2009). A customized and versatile high-density genotyping array for the mouse. Nat. Methods 6, 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadelaar S. M., Boesten L. S. M., Pires N. M. M., van Nieuwkoop A., Biessen E. A. L., Jukema W., Havekes L. M., van Vlijmen B. J. M., Willems van Dijk K. (2006). Local Cre-mediated gene recombination in vascular smooth muscle cells in mice. Transgenic Res. 15, 31–36 [DOI] [PubMed] [Google Scholar]