Advanced glycation end product (AGE) recognition by the receptor for AGEs (RAGE) (original) (raw)
. Author manuscript; available in PMC: 2012 May 10.
Published in final edited form as: Structure. 2011 May 11;19(5):722–732. doi: 10.1016/j.str.2011.02.013
SUMMARY
Nonenzymatic protein glycation results in the formation of advanced glycation end products (AGEs) that were implicated in the pathology of diabetes, chronic inflammation, Alzheimer’s disease, and cancer. AGEs mediate their effects primarily through a receptor-dependent pathway in which AGEs bind to a specific cell surface associated receptor, the Receptor for AGEs (RAGE). _N_ε-carboxy-methyl-lysine (CML) and _N_ε-carboxy-ethyl-lysine (CEL), constitute two of the major AGE structures found in tissue and blood plasma, and are physiological ligands of RAGE. The solution structure of a CEL containing peptide-RAGE V domain complex reveals that the carboxyethyl moiety fits inside a positively charged cavity of the V domain. Peptide backbone atoms make specific contacts with the V domain. The geometry of the bound CEL peptide is compatible with many CML (CEL) modified sites found in plasma proteins. The structure explains how such patterned ligands as CML (CEL)-proteins bind to RAGE and contribute to RAGE signaling.
Keywords: glycation, receptor for glycation end products, pattern recognition receptor, _N_ε-carboxy-methyl-lysine, _N_ε-carboxy-ethyl-lysine
The products of nonenzymatic glycation and oxidation of proteins, the early and the advanced glycation end products (AGEs), are a heterogeneous class of compounds that form under diverse circumstances in response to cellular stress(Brownlee et al., 1984). Specific AGE compounds, namely _N_ε-carboxy-methyl-lysine (CML) and _N_ε-carboxy-ethyl-lysine (CEL) (Figure 1A), due to their interactions with the Receptor for AGEs (RAGE), have been linked to complications of diabetes and chronic inflammation, the severity of Alzheimer’s disease, and cancer(Ishiguro et al., 2005; Ramasamy et al., 2005a; Ramasamy et al., 2005b; Thornalley, 1999; Valente et al.). RAGE is located in the major histocompatibility complex class III (MHC III) region suggesting its involvement in immune responses(Schmidt and Stern, 2001; Sugaya et al., 1994). RAGE is a member of the immunoglobulin superfamily of cell surface molecules and consists of three extracellular immunoglobulin domains, V, C1, and C2, a transmembrane helix and a short cytosolic tail(Hori et al., 1995; Neeper et al., 1992). In spite of their structural diversity, AGEs bind only to the V domain of RAGE(Kislinger et al., 1999; Xie et al., 2008). This binding does not accelerate clearance or degradation but rather begins a sustained period of cellular activation mediated by receptor-dependent signaling, leading to inflammation. It is proposed that RAGE activation is largely responsible for the pathogenicity associated with AGEs(Hofmann et al., 1999; Schmidt et al., 1995). Soluble RAGE (sRAGE), a variant containing extracellular V, C1, and C2 domains serves as a scavenger and is a potent inhibitor of RAGE signaling(Hori et al., 1995).
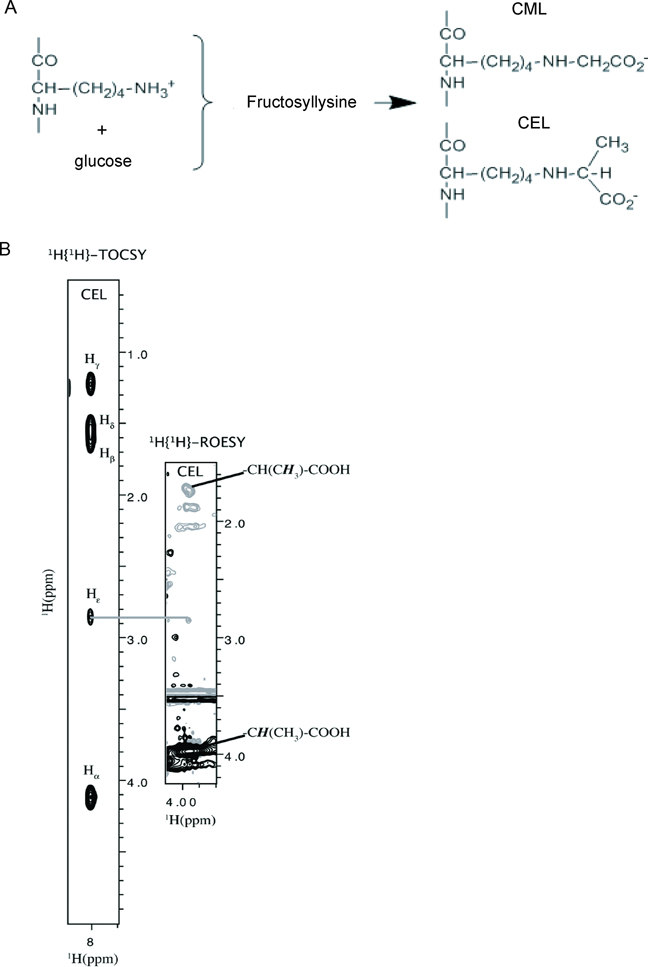
Figure 1.
CEL (CML) containing peptides can be produced by both glycation and chemical synthesis. (A) Fructosyllysine, CML and CEL are the products of early and advanced glycoxidation of sugars. (B) Synthetic CEL-peptide, DEF(CEL)ADE, contains a correct chemical structure of _N_ε-carboxy-ethyl-lysine. To characterize CEL-peptide, we collected two 2D homonuclear NMR experiments, 1H,1H TOCSY and 1H, 1H ROESY. The 1H, 1H TOCSY strip shows through bond correlation between the amide proton (8ppm) and side chain protons Hα (4.1 ppm), Hβ (1.7 ppm), Hγ (1.2 ppm), Hδ (1.5 ppm), and Hε (2.85 ppm) of CEL. The 1H, 1H ROESY strip shows through space correlations between Hε and carboxyethyl protons CH(CH3)-COOH (3.8 ppm) and CH(_CH_3)-COOH (1.7 ppm) of CEL, confirming the presence of a proper chemical structure of _N_ε-carboxy-ethyl-lysine. See also Figure S1.
Other endogenous ligands are implicated in amplifying RAGE dependent proinflammatory signaling: Cytokine-like mediators of the S100 family(Hofmann et al., 1999) and amphoterin(Taguchi et al., 2000), a nuclear protein released by necrotic cells. Unlike AGEs and possibly amphoterin that bind to a single domain, all three extracellular domains of RAGE are involved in binding S100 proteins: S100A12 binds to the C1 domain(Xie et al., 2007), S100B binds to V and C1(Dattilo et al., 2007; Ostendorp et al., 2007), and S100A6 binds to V and C2(Leclerc et al., 2007). The differences between RAGE binding sites may account for the diverse cellular responses caused by S100 proteins. Another major class of AGEs, imidazolones have also been implicated as possible ligands of RAGE(Thornalley, 1998). The structural diversity of RAGE ligands and the fact that RAGE recognizes a class of ligands, such as AGEs, led to the hypothesis that RAGE is a pattern recognition receptor(Chavakis et al., 2003).
Despite the fact that AGE-RAGE biology has been studied for more than 20 years, very little is known about the structural biology of AGE-RAGE complexes. This is mostly due to the extensive heterogeneity of AGEs created by glycation reactions: Glycation reactions are not largely dependent on sequence specificity, and lysine and arginine residues, which are particularly susceptible to glycation, are very common in proteins. The binding of AGE modified proteins to the V domain of RAGE depends weakly on either the primary or tertiary structure of the AGE modified sites. And while individual CML (CEL) structures bind to the V domain of RAGE weakly, constitutive oligomerization of RAGE provides a mechanism for increasing the number of binding sites and subsequently, the binding affinity of AGEs(Thornalley, 1998; Xie et al., 2008).
Many AGE structures are present in plasma at very low concentrations and thus, are unlikely to contribute significantly to RAGE signaling(Thornalley et al., 2003). At the same time, CML and CEL are major AGEs found in tissue and blood plasma and have been established as ligands of RAGE(Schmidt et al., 1996; Thornalley et al., 2003). Structural information is critical to understand the mechanisms of RAGE signaling and to design and optimize effective therapeutic agents against RAGE caused pathologies.
Here, we solved a solution structure of a CEL containing peptide V domain complex. Structure of the V domain is closely homologous to the variable domain of an antibody. CEL peptide binds to the surface formed by the C, F and G strands. The homologous surface in the variable domain of an antibody is used for heterodimerization. We showed that both the CEL moiety and the peptide backbone make specific contacts with the V domain of RAGE. The structure suggests how RAGE can binds to a large variety of CEL(CML) modified proteins. Specific binding of CML (CEL) modified proteins to RAGE suggests that the receptor functions as a sensor for cellular stress.
Results
A variety of extracellular proteins containing CML or CEL and possessing unrelated primary sequences bind to RAGE. Surprisingly, free CML and CEL do not bind to RAGE(Xie et al., 2008). Only CML or CEL embedded in a peptidic structure are capable of binding(Xie et al., 2008). Thus, to structurally characterize AGE-RAGE interactions, we synthesized short peptides containing CML and CEL (Figure 1B and Figure S1). The amino acid sequences of the peptides represent the primary glycation sites found in human serum albumin, HSA, a major plasma protein.
The affinities of CML (CEL) peptides for the V domain were estimated by monitoring changes in the native tryptophan fluorescence of the V domain upon ligand binding. Unmodified peptides, F(K)DLGEE and DEF(K)ADE, bind to the V domain with low affinities, whereas the binding of modified peptides is ~6–7-fold greater (Table 1 and Figure S2). As expected, the fluorescence titration indicated that the binding depends only slightly on the primary sequence of the peptide and even less so on the type of AGE modification. For example, the modified peptides F(CML)DLGEE and DEF(CML)ADE bind with 97 uM and 87 uM affinity, respectively, and F(CEL)DLGEE and DEF(CEL)ADE bind with 104 uM and 93 uM affinity, respectively. Overall, the binding affinities for all of the modified peptides examined were very similar to one another.
Table 1.
Binding affinities of the CML(CEL) peptides for the wild type and mutants of V domain.
| Peptidesequence | V domainKd, µM1 | R98A V domainKd, µM1 | K52A V domainKd, µM1 | K52A, R98A V domainKd, µM1 |
|---|---|---|---|---|
| DEF(CML)ADE | 97 ± 3 | 725 ± 17 | 528 ± 19 | 830 ± 27 |
| DEF(CEL)ADE | 104 ± 5 | 685 ± 26 | 560 ± 32 | 912 ± 42 |
| DEFKADE | 617 ± 24 | - | - | - |
| F(CML)DLGEE | 87 ± 5 | - | - | - |
| F(CEL)DLGEE | 93 ± 6 | - | - | - |
| FKDLGEE | 673 ± 38 | - | - | - |
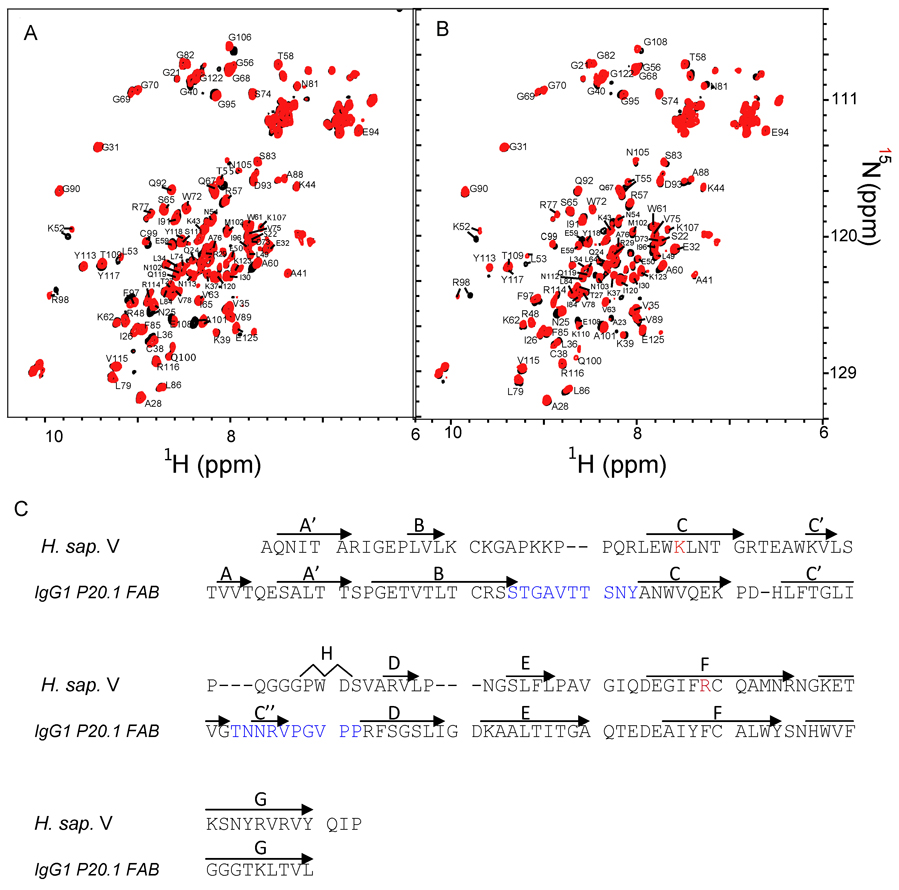
Since CML and CEL differ only by a methyl group (Figure 1 and Figure S1) we hypothesized that the CML and CEL peptides bind to the same molecular surface of the V domain. NMR chemical shifts of backbone amide protons and nitrogens are exquisitely sensitive to the changes in chemical environment induced by ligand binding. These changes allowed us to identify amino acid residues of the V domain that are affected by the binding of either DEF(CML)ADE (CML-PEP) or DEF(CEL)ADE (CEL-PEP) (Figure 2B,C and Figure S3). Titrating the peptides into the V domain led to gradual changes in the chemical shift of the residues Asn25, Lys52, Glu97, Arg98, Cys99, Ala101, and Lys110. This set of residues was virtually the same for CML-PEP and CEL-PEP suggesting that the V domain does not discriminate between these species and the same interaction surface is involved in CML and CEL binding.
Figure 2.
RAGE V domain does not discriminate between CML and CEL containing peptides. (A) Overlay of 15N-HSQC of free (black) and CML-PEP bound (red) V domain. (B) Overlay of 15N-HSQC of free (black) and CEL-PEP bound (red) V domain. (C) Sequence alignment of V domain and a V-type domain from a heavy chain antibody IgG1 P20.1. Amino acid residues involved in CML (CEL) binding are in red. CDR1 and CDR2, the hypervariable regions of IgG1 P20.1, are in blue. Secondary structure elements are shown above the sequences. See also Figure S3.
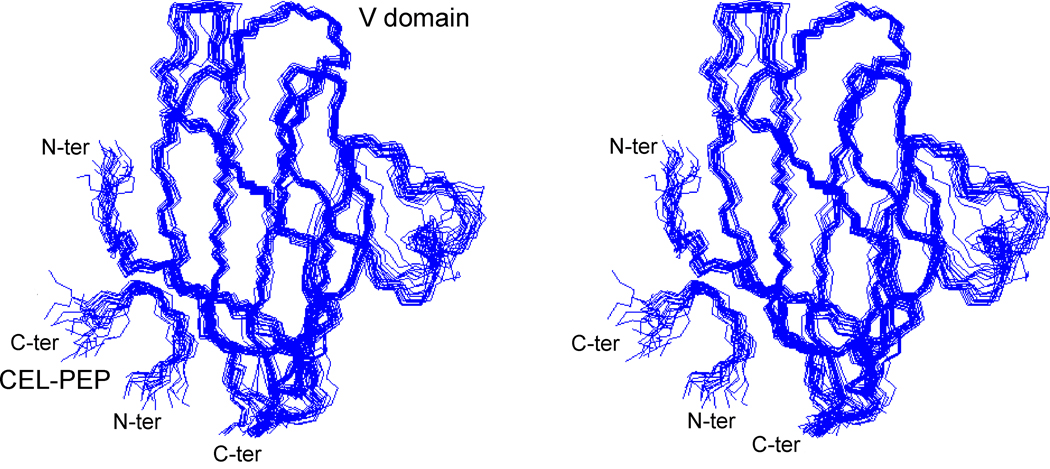
To elucidate the chemical nature of the interaction between the V domain of RAGE and CEL (CML) containing proteins, we studied the solution structure of the V domain (residues 23–123 of the 356-residue RAGE) in complex with the 7-residue CEL-PEP. The structured regions comprised residues 23–92 of the V domain and 3–5 of CEL-PEP. Based on the constraints obtained from NMR experiments, 25 structures with the lowest target function values were superimposed (Figure 3 and Table S1). The solution structure of the V domain-CEL-PEP complex is similar to that of the free V domain(Matsumoto et al., 2008) and V domain within a VC1 construct(Koch et al., 2010). The root mean square (r.m.s.) deviations between the solution structure of the complexed and free V domain (PDB code 2E5E) or V domain within VC1 construct (PDB code 3CJJ) are 1.8 Å and 1.4 Å for the backbone and 2.6 Å and 1.9 Å for all heavy atoms in of the ordered regions. The large loop between strands C’ and D was poorly defined in the free V domain(Matsumoto et al., 2008) and excluded from structural comparison.
Figure 3.
Stereoview of the overlay of 25 lowest energy CEL-PEP V domain backbone traces (PDB code 2L7U). N- and C-termini of the V domain and CEL-PEP are indicated. Figure is prepared by using Molmol (Koradi et al, 1996). See also Table S1.
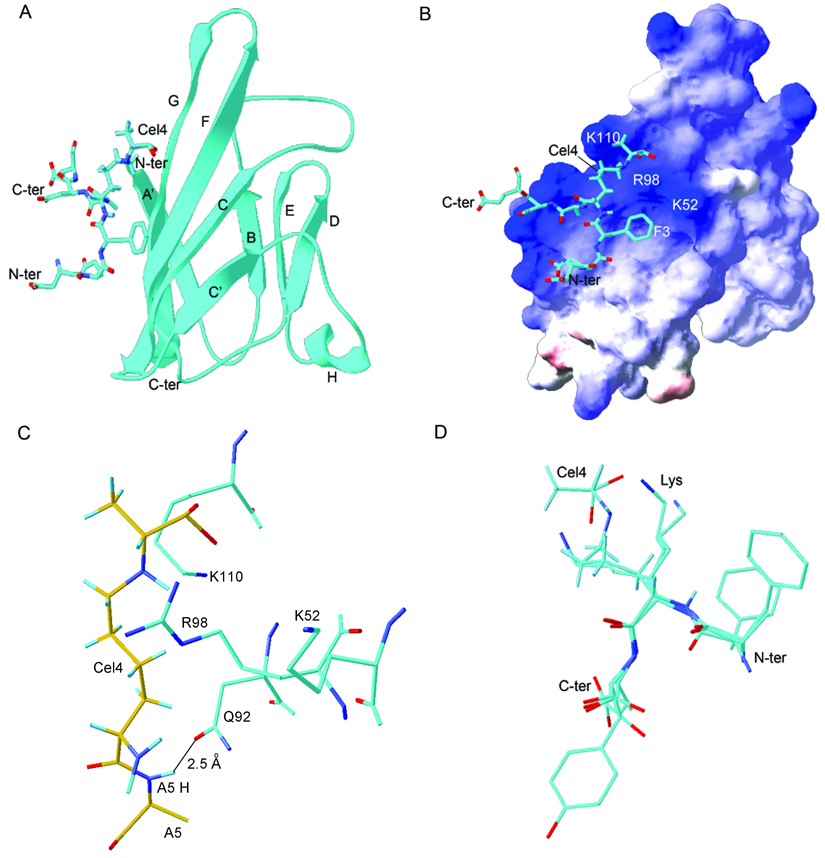
The solution structure of the V domain (Figure 4A) closely resembles the structure of conventional immunoglobulin V-type domains. Secondary structure elements were labeled following the immunoglobulin convention(Bork et al., 1994; Chothia and Jones, 1997). The network of hydrogen bonds in the RAGE V domain is different from that of the immunoglobulin-like domains found in cell adhesion receptor molecules, such as ICAM and JAM, suggesting a different mode of action. The closest structural analog is a variable domain of the IgG1 P20.1 FAB light chain (PDB code 2ZPK(Nogi et al., 2008)) (Figure 2C, Figure 4A and Figure S4).
Figure 4.
Solution structure of the CEL-PEP-V domain complex. (A) Structure of CEL-PEP bound to V domain. V domain is shown in ribbon representation. Elements of secondary structure are labeled following the immunoglobulin convention(Bork et al., 1994). (B) Electrostatic potential is mapped onto the molecular surface of the V domain. Positively and negatively charged surfaces are indicated in blue and red, respectively. (C) V domain amino acid residues located within 5 Å from the CEL moiety of CEL-PEP. Putative hydrogen bond between the backbone amide proton of Ala5 and the side chain carbonyl group of Asn112 is indicated. Carbon atoms of CEL-PEP and V domain are in yellow and cyan, respectively. (D) Structural alignment of CEL-PEP with three short segments from human serum albumin (HSA) containing lysines(Wa et al., 2007), 11-FKD, 56-AKT, and 261-AKY. The lysines in these sequences were shown to be glycated under elevated concentrations of D-glucose. See also Figure S4 and Table S2.
The V domain structure consists of seven-strands connected by six loops, to form two beta-sheets. The disulfide bond between Cys38 and Cys99 links these sheets into a beta-sandwich structure. Two anti-parallel beta strands, B and D, are shorter than in the conventional, IgG1 P20.1 FAB V-type domain(Bork et al., 1994) (Figure 2C). This arrangement allows loops BC and C’D to be packed against the core of the protein rather than protruding into the solvent. Loops BC and C’D contain the hypervariable CDR1 and CDR2 regions of the antibody that are involved in antigen binding (Figure 2C and Figure S4). Two other regions distinct from conventional immunoglobulin structures are a helix between strands C’ and D and the apparent lack of the hydrogen bonds necessary to form the C” strand conventionally found in variable-type domains.
The electrostatic potential mapped onto the molecular surface of the V domain is shown in Figure 4B. There is only one obvious hydrophobic cavity close to the C terminus formed by Ile 30, Pro 87, Ala 88, Ile 91, Tyr 118 around loop A’B and loop EF. The electrostatic surface potential of the V domain reveals that the molecular surface is covered by positive charges. In particular, there are two areas where positive charges are densely localized to form a cationic center. The first one consists of Lys52, Arg98, and Lys110 and is positioned on a relatively flat molecular surface. IgG1 utilizes this area to form heterodimers between light and heavy chain variable domains (Figure 4A and Figure S4). The second area of positive charge consists of Lys43, Lys44 on loop BC and Arg104 on loop FG. Conventional antibodies utilize the latter surface for specific ligand binding(Bork et al., 1994;,Nogi et al., 2008).
The binding conformation of the CEL-PEP peptide backbone is bent at CEL and forms a loop possibly due to steric hindrance caused by the bulky CEL group (Figure 3 and Figure 4). The CEL-PEP binding site is small, spanning 180 Å2 and sits on a positively charged groove formed by the C, F and G strands. Conventional antibodies use this surface for heterodimerization (Figure 4A and Figure S4). The binding site for CEL-PEP is relatively flat which probably reflects the lack of specificity for primary structure observed in binding experiments.
The CEL moiety of CEL-PEP makes the majority of intermolecular contacts (Figure 4C and Table S2). CEL-PEP also contacts only a limited number of residues in the V domain of RAGE (Figure 4C and Table S2). Ionic interactions predominate CEL-PEP-V domain interactions since an increase in ionic strength from 200 mM to 400 mM abolished CEL-PEP binding. We detected an extended network of nOes confidently placing the negatively charged carboxyethyl head group of CEL into the groove formed by positively charged Lys52, Lys110, and Arg98 (Figure 4C, Table S2 and Figure S4C). The rest of CEL fits snugly into a groove formed by the F and G strands, which may provide additional hydrophobic contacts. To further demonstrate that Lys52 and Arg98 play a major part in the binding of CML (CEL) containing peptides to the V domain, we made single R98A and K52A, and double R98A, K52A mutants of the V domain. The binding affinities of CML-PEP and CEL-PEP for the mutant V domains are 5–10-fold weaker than for the wild type V domain, and comparable to that of unmodified PEP (Table 1 and Figure S2).
CEL moiety binds to the V domain in an extended conformation that is longer than any of the amino acids side chains. As a result, substituting a negatively charged side chain such as Glu for CEL will lead to loss of binding. Importantly, the methyl group of CEL is oriented away from the binding site suggesting that this group does not contribute to the binding free energy. This configuration of CEL would explain why CML, which lacks the methyl group, has a very similar binding affinity and fits into a very similar interaction surface. Consistent with our previous binding experiments, an early glycation end product, fructosyllysine (Figure 1A), which possesses a bulky head group, will not fit into the CEL binding site on the V domain (Xie et al., 2008).
Our earlier observations suggest that interactions between the CEL moiety and V domain are not sufficient to provide stable binding since neither free CEL nor CML stably bind to the V domain(Xie et al., 2008). We hypothesized that the peptide backbone conformation is important for CEL-PEP binding to the V domain. Indeed the backbone amide proton of CEL-PEP Ala5 is located within 2.5 Å of the V domain Asn112 side chain carbonyl group and possesses geometry consistent with a hydrogen bond (Figure 4C). The chemical shift of the Ala5 amide proton exhibits a downfield shift from 7.6 ppm to 8.2 ppm upon CEL-PEP binding to the V domain, which is also indicative of hydrogen bond formation. We detected intermolecular contacts between Hβ of the Ala5 side chain and Hδ of Arg114, which may strengthen the CEL-PEP peptide backbone-V domain interaction (Table S2 and Figure S4C). Additional hydrophobic contacts are supplied by V domain residues Trp61, Ile96 and Phe97, which together extend the hydrophobic surface along face of Phe3, since we detected an intermolecular nOe between Hβ of Phe3 and Hδ of Ile96 (Figure S4C). At the same time, the interaction between the benzene ring of Phe3 and the V domain is not specific since we did not identify extensive network of nOes from the V domain to Phe3 benzene ring (Figure 4C and Table S2). The side chain of Phe3 is not well defined in the V domain-CEL-PEP complex (Figure S4D). In addition, during the CEL-PEP-V domain titration residues Trp61, Ile96 and Phe97 did not undergo the large chemical shift changes expected for ring current shifts (Perkins et al., 1979). This hydrophobic surface is large enough to accommodate any side chain, possibly providing an increase in binding affinity without a subsequent increase in specificity.
To rationalize the ability of RAGE to bind various CML and CEL modified proteins, we structurally aligned CEL-PEP with three-dimensional structures of loops from BSA reported to contain major glycation sites(Wa et al., 2007) (Figure 4D). Three out of six loops possessed conformations that are compatible with binding to the V domain. Importantly, the conformation of the backbone amide group that follows the AGE modified lysines in these loops are almost identical to that of CEL-PEP. Moreover, it is known that glycation disrupts local secondary structure rendering the area immediately around the modified site extremely flexible (Povey et al., 2008). This suggests that even CML (CEL) modified sites, which do not initially possess proper binding geometry, may be able to bind to the V domain by using induced fit.
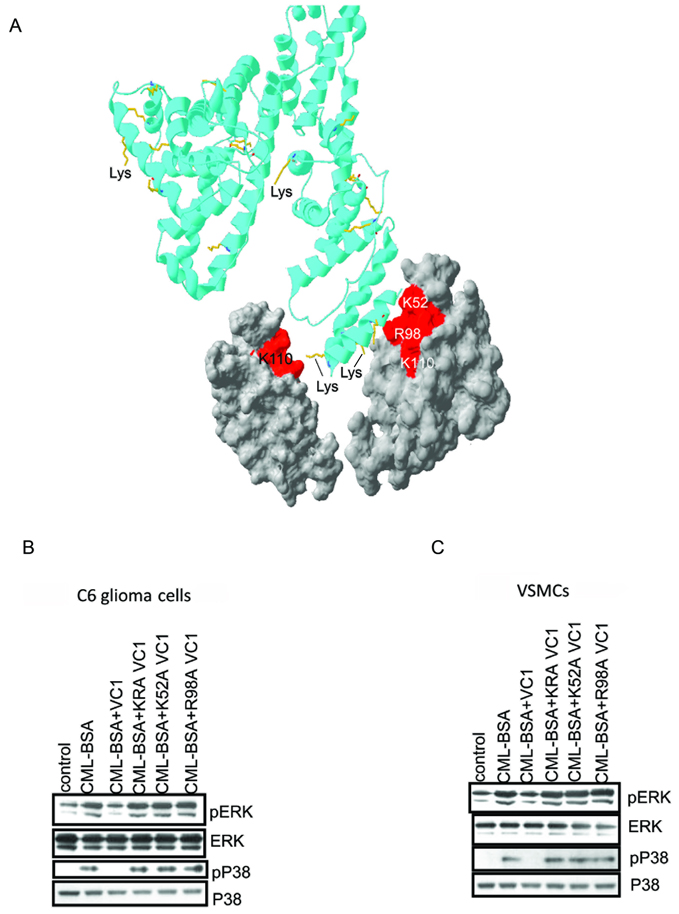
Soluble RAGE (sRAGE), works as a scavenger to remove ligands capable of activating RAGE expressing cells(Hofmann et al., 1999). We used this function of sRAGE to test whether our structure of the CEL-PEP-V domain reflects physiological interactions between RAGE and CML modified proteins. Instead of the V domain that binds to CML peptides only weakly, we used the VC1 domain of RAGE that dimerizes(Xie et al., 2007; Koch et al., 2010; Zong et al., 2010) (Figure 5A and Figure S5A) and thus, is capable of binding multiple CMLs with increased binding affinity. CML modified BSA was used as a RAGE activating ligand. To interrogate the functional implications of these findings, we used two RAGE-expressing cell types, C6 glioma cells and primary murine aortic vascular smooth muscle cells (VSMC) (Brett et al., 1993; Taguchi et al., 2000). Cellular activation was monitored by observing the increase in phosphorylation levels of two key signal transduction proteins previously shown to be downstream of RAGE: ERK and P38 (MAPK)(Kislinger et al., 1999).
Figure 5.
Mutants of the VC1 domains of RAGE fail to suppress CML-BSA induced RAGE signaling. (A) Cartoon model of how RAGE dimerization promotes V domain binding to multiple CML moieties on CML-BSA (ribbon). The molecular surface of the V domain involved in CML(CEL) binding is in red. Only two V domains are shown. Lysines of BSA, which may undergo glycation are in yellow. The cartoon model was prepared by using SWISS-PDB Viewer (Guex et al, 1997). (B, C) Single K52A and R98A, and double K52A, R98A (KRA) mutants of the VC1 domains do not interfere with CML-BSA induced RAGE signaling in both C6 rat glioma (b) and mouse VSMC cells (c). See also Figure S5.
Wild type VC1 inhibited CML-BSA induced RAGE signaling in both C6 glioma cells and VSMC cells, as evidenced by no increase in the levels of phosphorylated ERK (pERK) and phosphorylated MAPK (pP38) relative to the control lane (Figure 5B,C and Figure S5B,C). Total ERK and MAPK levels remained constant for all experiments. The two single mutants, R98A and K52A, and one double mutant of VC1, R98A, K52A (KRA), failed to inhibit RAGE signaling induced by CML-BSA, strongly suggesting that the identified CEL (CML) binding site is critical for the CML-BSA induced activation of RAGE.
Discussion
The structure of the V domain with its physiological ligand, CEL-PEP, revealed how RAGE is able to recognize CML(CEL) modified proteins when the modification site can possess very different primary, secondary, and tertiary structures. The V domain of RAGE makes molecular contacts with both the CEL(CML) moiety and the peptide backbone in the immediate vicinity of CEL(CML). The V domain does not discriminate between CEL and CML. The negative charge of CEL (CML) is a critical determinant for binding to the positively charged V domain surface. The extended geometry of CEL(CML) and the distance from the negative carboxyl group to the peptide backbone is also critical for molecular recognition. These two conditions will preclude binding of either negatively charged amino acids or bulky early glycation products, such as fructosyllysine, to the CEL-PEP binding site. The torsional angles of the CEL-PEP peptide backbone when bound to the V domain, allow different sequences within the AGE modified protein to fit into the binding site. The latter explains why RAGE can bind to various CML (CEL) modified proteins found in tissue and in plasma.
Binding of CEL-PEP to the V domain is clearly specific. Is the binding of CML(CEL) proteins to RAGE physiologically important and how does it relate to the immune response caused by RAGE signaling? CML and CEL modified proteins are found under normal physiology(Thornalley et al., 2003). However, their concentration is low and individual proteins have at most only a single CML (CEL) modified lysine. Since binding of a single CML peptide to the V domain is weak this event will not trigger RAGE signaling(Xie et al., 2008). Under cellular stress, caused by different pathologies including diabetes, Alzheimer’s disease, or cancer, the concentration of CML (CEL) proteins increases, thus increasing the amount of multiple-modified proteins(Brownlee, 1995). Since RAGE is a constitutive oligomer(Xie et al., 2007; Koch et al., 2010; Zong et al., 2010), polyvalent engagement of CML-proteins and RAGE results in tight binding, thus, triggering the RAGE-dependent immune response. This mechanism suggests a physiological role for RAGE as a sensor of cellular stress caused by various pathologies(Schmidt et al., 2000). It also highlights an opportunity to interfere with RAGE signaling by obstructing low affinity ligand binding. The structure of the CML-PEP-V domain will rationalize these efforts.
Experimental Procedures
1) Reagents and Chemicals
Restriction enzymes and Taq polymerase were from NEB. All other chemicals used were reagent grade or better.
2) Solid phase CML (CEL)-peptides synthesis
CML- and CEL-containing peptides were synthesized on the multiple synthesizer SYRO2000 (MultiSynTech GmbH, Witten, Germany) by using 9-fluorenylmethoxycarbonyl/_tert_-butyl (Fmoc/tBu)-chemistry on the Wang resin(Atherton et al., 1978) utilizing a global post synthetic alkylation approach. The side chains of Asp and Glu were protected with tBu and the side chain of Lys with 4-methyltrityl (Mtt). Fmoc-amino acid derivatives were activated with N,N’-diisopropylcarbodiimide (DIC) and N-hydroxy-benzotriazole (HOBt) at equimolar ratios, added to the resin with eight molar excess, and reacted at room temperature (RT) for 70 min. After completion of the synthesis, the Fmoc-group was cleaved with piperidine and the free N-terminus was protected again with a mixture of di-_tert_-butyl dicarbonate (Boc2O, 20 eq) and N,N-diisopropylethylamine (DIPEA, 10 eq) in N,N-dimethylformamide (DMF) at RT overnight. The Mtt-group was cleaved with 2% (v/v) trifluoroacetic acid (TFA) and 1% (v/v) triisopropylsilane (TIS) in dichloromethane (DCM) two times for 30 min. The resin was washed three times with DCM and dried before the unprotected ε-amino groups of the lysine residues were alkylated with 2-chlorotrityl 3-bromopropanoate (or 2-chlorotrityl bromoacetate) (20 eq) in the presence of DIPEA (10 eq) in DCM to obtain the protected CEL- (or CML-) peptides, respectively. Peptides were cleaved with TFA containing 12.5% (v:v) of a scavenger mixture (ethandithiole, m-cresole, thioanisole and water, 1:2:2:2) at RT for 2 hrs. The peptides were precipitated with cold diethyl ether and purified by RP-HPLC with a linear aqueous acetonitrile gradient (5 mmol ammonium formate, pH 3.2) as ion pair reagent by using a Jupiter C18-column (21.2 mm internal diameter, 250 mm length, 15 µm particle size, 30 nm pore size) (Phenomenex Inc., Torrance, USA). The purities of the peptides were confirmed by RP-HPLC and the molecular weight by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS; 4700 proteomic analyzer; Applied Biosystems GmbH, Darmstadt, Germany) using α-cyano-4-hydroxycinnamic acid matrix.
3) Preparation of 2-Chloro-trityl 3-bromopropanoate and 2-chloro-trityl 3-bromoacetate
3-Bromopropionic acid (9,6 mmol, 0,86 mL) or bromoacetic acid (9,6 mmol, 3,0 g) were mixed with equimolar amounts of 2-chlorotrityl chloride (1.34 g) and DIPEA (1.65 mL) in dichloromethane (25 mL). This mixture was stirred at RT for 1 h before the solvent was removed on a rotary evaporator. The dry products were used without further purification to alkylate the side chains of lysine residues.
4) Plasmid construction
Human RAGE cDNA library clone BC020669 was obtained from Open Biosystems and used as a template for PCR amplifications. DNA coding for the V domain (amino acids 24–125) was PCR-amplified using Taq polymerase and oligonucleotides 5’-TTTCATATGGCTCAAAACATCACAGCCCGGATTGG and 3’-TTTGTCGACTCATTCTGGCTTCCCAGGAATCTG containing flanking 5'-_Nde_I and 3'-_Sal_I restriction sites. The restriction-digested PCR products were ligated into expression vector pET28a (Novagen), which confers kanamycin resistance. The resulting plasmid, pET28-V, expresses a C-terminal His-tagged V domain of RAGE. DNA coding for VC1 fragments (amino acids 23–243) were PCR amplified using oligonucleotides 5’-TTTCATATGGCTCAAAACATCACAGCCCGGATTGG and 3’-TTTGAGCTCCACCACCAATTGGACCTCCTCCAG containing 5’-_Nde_I and 3’-_Xho_I restriction sites. DNA fragments were subcloned into the _Nde_I and _Xho_I sites of pET15b vector (Novagen), which confers ampicillin resistance. The resulting plasmid, pET15b-VC1, expresses an N terminal His-tagged VC1 domain.
5) Labeling, Expression, and Purification of Wild Type and Mutant V Domains
To uniformly label the V domain of RAGE, pET28-V, pET28-R98A-V, pET28-K52A-V or pET28-K52A-R98A-V were transformed into E. coli strain BL21(DE3) Codon+ (Novagen). For U-15N labeling, cells were grown at 37 °C in minimal medium (M9) containing 35 mg/liter kanamycin and 1 g/liter [15N]ammonium chloride as the sole nitrogen source. For U-13C,15N labeling, cells were grown at 37 °C in M9 medium containing 35 mg/liter kanamycin, 1 g/liter [15N]ammonium chloride, and 2 g/liter [13C]glucose instead of unlabeled glucose as the sole carbon source. Cells were grown to 0.7 _A_600 at 37 °C, induced with 0.5 mm isopropyl 1-thio-b-d-galactopyranoside (IPTG), and grown overnight. Cells were harvested and resuspended in 20 mM Hepes-Na [pH 7.0] buffer, containing 8 M urea and heat lysed at 100 °C for 10 min. The lysate was centrifuged, and the supernatant was loaded onto a nickel-nitrilotri-acetic acid-agarose (Ni-NTA) column (Qiagen). The column was washed with 20 mM Hepes-Na buffer, pH 7.0, and the protein was allowed to re-nature on the column before eluting with 20 mM Hepes-Na, pH 7.0, containing 500 mM imidazole. Fractions containing the eluted protein were pooled and dialyzed into NMR buffer (10 mm sodium phosphate, pH 6.5, 100 mm NaCl, 0.02% (w/v) NaN3). The C-terminal His tag of the V domain was cleaved by thrombin (Novagen) at room temperature for 1 h before gel filtration chromatography on a SE-75 column (Amersham Biosciences). The fractions containing the eluted protein were concentrated by using Ultra-Centricones (Millipore). Purity was estimated to be >95% by Coomassie-stained SDS-PAGE.
6) Expression and Purification of Wild Type and Mutant VC1 Domains
RAGE fragments were overexpressed in E. coli strain OrigamiB-(DE3) (Novagen), grown at 37 °C to OD600 ~0.8, adjusted to 20 °C for 30 min, induced with 1 mM IPTG, and allowed to express for 4–6 hours. Cells were lysed at 4 °C in lysis buffer (20 mM Tris-HCl [pH 8.0], 20 mM imidazole, 300 mM NaCl) in the presence of lysozyme (5 mg/mL), followed by sonication (5 min with a 50% duty cycle). Clarified lysate was initially purified on a Ni-NTA (Qiagen) column equilibrated with lysis buffer and eluted with 20 mM Tris-HCl [pH 8.0], 100 mM imidazole, 300 mM NaCl. The C-terminal His tag of the VC1 domain was cleaved by thrombin (Novagen) at room temperature for 1 h before gel filtration chromatography on a SE-75 column (Amersham Biosciences). The fractions containing the eluted protein were concentrated by using Amicon-Ultra-Centricones (Millipore). Residual endotoxin was removed from the sample by repetitive use of EndoTrap Red (Lonza). The endotoxin level of the final protein solution was determined by using Gel Clot LAL Assay (Lonza) to be less than 0.1 EU/mL. Purity was estimated to be >95% by Coomassie-stained SDS-PAGE.
7) Preparation of CML-Bovine Serum Albumin (BSA)
We followed the protocol (Kislinger et al., 1999) that leads exclusively to CML modifications of lysine. Briefly, CML-BSA was prepared by incubating 5 mM BSA (fraction V, fatty acid free, endotoxin free bovine serum albumin, EMD, Gibbstown, NJ) in 150 mM sodium phosphate buffer [pH 7.4], containing 25 mM glyoxylic acid and 75 mM NaBH3CN, at 50 °C for 48 h. The reaction mixture was dialyzed against 10 mM Sodium phosphate buffer [pH 7.0] and 100 mM NaCl to remove unreacted glyoxylic acid and NaBH3CN, and stored at −20 °C in 20% glycerol. The extent of chemical modification was determined colorimetrically by using 2,4,6-trinitrobenzenesulfonic acid to measure the difference spectrum between lysine residues of modified and unmodified protein preparations(Habeeb et al., 1966). The extent of lysine modification of CML-BSA preparations was 21%.
8) Site-Directed Mutagenesis of the V and VC1 domains
To singly or doubly mutate the V domain of RAGE, the QuikChange II XL Site-Directed Mutagenesis Kit (Strategene) was used. Following mutagenic PCR, pET28-V was restriction digested with _Dpn_I for 1 hour and transformed into E. coli strain DH10B. Mutated plasmids were isolated and purified using Mini-Prep Kit (Qiagen). DNA sequencing identified plasmids pET28-R98A-V, pET28-K52A-V or pET28-K52A-R98A-V, which code for the appropriate mutant V domain.
To singly or doubly mutate the VC1 domain of RAGE, High-Fidelity DNA polymerase (New England BioLabs) was used. Following mutagenic PCR, pET15-VC1 was transformed into E. coli strain XL-1 Blue. Mutated plasmids were isolated and purified using Mini-Prep Kit (Qiagen). DNA sequencing identified plasmids pET15-R98A-VC1, pET15-K52A-VC1 and pET15-K52A-R98A-VC1, which code for the appropriate mutant VC1 domain.
9) NMR Experiments
CEL and CML containing peptides were assigned using 2D 1H,1H TOCSY and 1H,1H ROESY experiments (Cavanagh et al., 1996), which provide through bond and through space proton connectivities. Protein samples of the uniformly labeled, [_U_-13C,15N] and [_U_-15N], V domain, with concentrations ranging from 60 to 300 uM were dissolved in NMR buffer (10 mM potassium phosphate [pH 6.5], 100 mM NaCl, 0.02% (w/v) NaN3, in 90%H2O/10%D2O) and unlabeled peptide, CEL-PEP or CML-PEP, was added to a 1.2 molar excess. To obtain backbone resonance assignments of [_U_-13C,15N] V domain-CEL-PEP complex, standard triple resonance spectra 1H,15N HSQC, HN(CA)CO, HNCO, HN(CO)CA, HNCA, CBCA(CO)NH, and HNCACB(Cavanagh et al., 1996) were acquired at 298 K using an Avance Bruker spectrometer operating at a 1H frequency of 700 MHz equipped with a single Z-axis gradient cryoprobe. To obtain the side-chain resonance assignments of V domain bound to CEL-PEP, 1H, 13C HSQC, 1H, 13C 3D NOESY-HSQC and 3D HCCH-TOCSY experiments(Cavanagh et al., 1996) were acquired. To assign intramolecular nOes in CEL-PEP, a 15N-filtered 13C filtered NOESY-HSQC(Cavanagh et al., 1996; Iwahara et al., 2001; Zwahlen et al., 1997) was acquired on an NMR sample containing 300 uM [_U_-13C,15N] V domain and 200 uM unlabeled CEL-PEP, and a 13C-double filtered NOESY-HSQC(Cavanagh et al., 1996; Iwahara et al., 2001; Zwahlen et al., 1997) was acquired on the NMR sample with 350 uM [_U_-13C,15N] V domain and 200 uM unlabeled CEL-PEP. To assign intermolecular nOes, a 15N-edited 13C filtered NOESY-HSQC(Iwahara et al., 2001; Zwahlen et al., 1997) was acquired on an NMR sample containing 320 uM [_U_-13C,15N] V domain and 1mM uM unlabeled CEL-PEP, and a 13C-edited, 13C filtered NOESY-HSQC(Iwahara et al., 2001; Zwahlen et al., 1997) was acquired on the NMR sample with 300 uM [_U_-13C,15N] V domain and 1 mM unlabeled CEL-PEP. To identify the peaks from intramolecular 13C-bound protons, the NOESY spectra were acquired both with and without heteronuclear 13C decoupling during the indirect proton acquisition period. Peaks that were split in the absence of decoupling were assigned to intramolecular V domain nOes. All spectra were processed using TOPSPIN 2.1 (Bruker, Inc), and assignments were made using CARA(Masse and Keller, 2005).
To obtain translational diffusion coefficients, D, gradient diffusion experiments were performed using the pulse sequence described by Ferrage et al(Ferrage et al., 2003). A 100 uM protein sample of the uniformly labeled [_U_-15N] VC1 construct was dissolved in NMR buffer (10 mM potassium phosphate [pH 7.2], 100 mM NaCl, 0.02% (w/v) NaN3, in 90%H2O/10%D2O). The attenuation of the NMR signal due to the increase in the strength of the gradient field was used to calculate translational diffusion coefficients which, for a spherical Brownian particle is inversely proportional to the Stokes’ radius(Schimmel and Cantor, 1980),
KBT being the thermal energy, η the viscosity of the solvent and ra the Stokes’ radius. In our experiment the diffusion delay was Δ + 6τ = 1 s; each sine-shaped encoding gradient lasted δ = 1.3 ms. Translational diffusion coefficients were calculated by fitting integrated amide signal intensities using the equation:
where
γ is the proton gyromagnetic ratio, s the shape of the encoding and decoding gradient pulses and G their peak amplitude.
10) Structure Calculation
Structural calculations were carried out with Cyana 2.1(Guntert, 2004) using 986 distance restraints derived from 13C-edited NOESY and 15N-edited NOESY spectra, 150 pairs of backbone torsion angle restraints derived from TALOS(Cornilescu et al., 1999), 38 restraints for hydrogen bonds, and the restraints from one disulfide bond between Cys38 and Cys99. nOes were converted to upper limit distances using the CALIBA module in CYANA(Guntert, 2004). The reference volume determined by CALIBA was increased 2 times before conversion in order to loosen the distance restraints. All upper limit distances for intermolecular nOes were set to 6 Å. The geometry of unnatural amino acids CML and CEL were added to the CYANA library. Backbone torsion angle restraints for CEL-PEP were estimated by using the TALOS(Cornilescu et al., 1999). These experimental restraints are summarized in Table S1. To perform CYANA calculations, a single polypeptide chain was constructed for the V domain and CEL-PEP molecules.
Refinement: The CYANA-generated distance and angle restraints were converted into CNS format in CCPN(Fogh et al., 2002). The structure for the non-standard amino acids CEL and CML was generated and energy-minimized in PRODRG2(Schuttelkopf and van Aalten, 2004). A total of 1,000 structures were calculated, and the 200 lowest energy structures were subjected to water refinement and further analysis by PROCHECK_NMR(Laskowski et al., 1996). 80.5% of the V domain residues were in the most favorable regions of Ramachandran plot, 18.2% were in the additional allowed regions and 1.3% were in generously allowed regions. There were no residues in the disallowed regions of the Ramachandran plot. The structural statistics of the 25 best structures are reported in Table S1.
11) Fluorescence Titration
Measurements were performed on a Fluorolog-3 fluorescence spectrophotometer (HORIBA Jobin Yvon) at 25 °C in a 1-ml stirred cuvette. For fluorescence titration experiments, 100 nM of V domain dissolved in 10 mM phosphate buffer [pH 6.5] and 100 mM NaCl was used, and 1 mM solution of CML and CEL containing peptides was used to increase the concentration in 10 nM steps. Titrations in the absence of V domain and in the absence CML (CEL) peptides were performed as references. Tryptophan fluorescence was measured using an excitation wavelength of 280 nm. The fluorescence emission signal was subtracted from the signal of the reference titrations, and the differences adjusted by the dilution factor were plotted against the final concentration of added CML (CEL) peptide. Curve fitting (OriginLab) was performed to find the best values for Kd using a single site binding isotherm approximation (Eftink et al, 1997).
12) Cell Lines and Materials
Wild-type mice primary vascular smooth muscle cells were isolated and cultured from aortas and employed through passage 5 to 7. Rat C6 glioma cells were obtained from ATCC (CCL-107) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). Primary vascular smooth muscle cells and C6 glioma cells were seeded at 1 × 106 cells/100-mm dish in complete media and grown for 24 h before starvation overnight in serum-free media. After overnight starvation cells were stimulated with 10 mg/ml CML-BSA or preincubated (2 hrs at 4 °C) CML-BSA with at least equimolar amounts of single mutants K52A, R98A, double mutant K52A-R98A, or wild type VC1 domain for the indicated time point, rinsed with ice-cold phosphate-buffered saline, and lysed using lysis buffer (Cell Signaling Technology) containing 1 mM phenylmethylsulfonyl fluoride and Complete Protease Inhibitors (Roche Applied Science).
13) Western blot analysis
Total cell lysate was immunoblotted and probed with ERK, p-ERK, P-38 and pP-38 specific antibody (Cell Signaling Technology), HRP-conjugated donkey anti-rabbit IgG (Amersham Pharmacia) or HRP-conjugated sheep anti-mouse IgG (Amersham Pharmacia) were used to visualize the bands on the gel. After probing with the p-ERK and pP-38 antibodies, membranes were stripped of bound immunoglobulins and reprobed with ERK or p38 antibody for relative total protein. Blots were scanned with an AlfaImager TM 2200 scanner with AlfaEase (AlfaImager) FC 2200 software. Results are reported as a relative of test antigen to relative total proteins. In all Western blot studies, at least 3 cell lysates per group were used; results of representative experiments are shown.
Supplementary Material
01
Acknowledgements
This work was supported by funding from the American Diabetes Association 1-06-CD-23 and the National Institutes of Health R01 GM085006-01A2. We thank Gregory McLaughlin (University at Albany) for his help in preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers.
Coordinates and chemical shift assignments of the CEL-PEP V domain complex have been deposited in the Protein Data Bank with accession number 2L7U.
Supplemental Information.
Supplemental Information includes five figures and two tables.
References
- Atherton E, Fox H, Harkiss D, Sheppard RC. J. Chem. Soc., Chem Commun. 1978:537–539. [Google Scholar]
- Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- Brett J, Schmidt AM, Zou YS, Yan SD, Weidman E, Pinsky D, Neeper M, Przysiecki M, shaw A, Migheli A, Stern D. Tissue distribution of the receptor for advanced glycation end products (RAGE): expression in smooth muscle, cardiac myocyte, and neural tissue in addition to vasculature. Am. J. Pathol. 1993;143:1699–1712. [PMC free article] [PubMed] [Google Scholar]
- Brownlee M. Advanced protein glycosylation in diabetes and aging. Annu Rev Med. 1995;46:223–234. doi: 10.1146/annurev.med.46.1.223. [DOI] [PubMed] [Google Scholar]
- Brownlee M, Vlassara H, Cerami A. Nonenzymatic glycosylation and the pathogenesis of diabetic complications. Ann Intern Med. 1984;101:527–537. doi: 10.7326/0003-4819-101-4-527. [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy: Principles and practice. San Diego: Academic Press; 1996. [Google Scholar]
- Chavakis T, Bierhaus A, Al-Fakhri N, Schneider D, Witte S, Linn T, Nagashima M, Morser J, Arnold B, Preissner KT, Nawroth PP. The pattern recognition receptor (RAGE) is a counterreceptor for leukocyte integrins: a novel pathway for inflammatory cell recruitment. J Exp Med. 2003;198:1507–1515. doi: 10.1084/jem.20030800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Jones EY. The molecular structure of cell adhesion molecules. Annu Rev Biochem. 1997;66:823–862. doi: 10.1146/annurev.biochem.66.1.823. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Dattilo BM, Fritz G, Leclerc E, Kooi CW, Heizmann CW, Chazin WJ. The extracellular region of the receptor for advanced glycation end products is composed of two independent structural units. Biochemistry. 2007;46:6957–6970. doi: 10.1021/bi7003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftink MR. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 1997;278:221–257. doi: 10.1016/s0076-6879(97)78013-3. [DOI] [PubMed] [Google Scholar]
- Ferrage F, Zoonens M, Warschawski DE, Popot JL, Bodenhausen G. Slow diffusion of macromolecular assemblies by a new pulsed field gradient NMR method. J Am Chem Soc. 2003;125:2541–2545. doi: 10.1021/ja0211407. [DOI] [PubMed] [Google Scholar]
- Fogh R, Ionides J, Ulrich E, Boucher W, Vranken W, Linge JP, Habeck M, Rieping W, Bhat TN, Westbrook J, et al. The CCPN project: an interim report on a data model for the NMR community. Nat Struct Biol. 2002;9:416–418. doi: 10.1038/nsb0602-416. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Habeeb AF. Determination of free amino groups in proteins by trinitrobenzenesulfonic acid. Anal Biochem. 1966;14:328–336. doi: 10.1016/0003-2697(66)90275-2. [DOI] [PubMed] [Google Scholar]
- Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, Nagashima M, Lundh ER, Vijay S, Nitecki D, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Nakaigawa N, Miyoshi Y, Fujinami K, Kubota Y, Uemura H. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate. 2005;64:92–100. doi: 10.1002/pros.20219. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Wojciak JM, Clubb RT. Improved NMR spectra of a protein-DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered NOESY experiment. J Biomol NMR. 2001;19:231–241. doi: 10.1023/a:1011296112710. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Fu C, Huber B, Qu W, Taguchi A, Du Yan S, Hofmann M, Yan SF, Pischetsrieder M, Stern D, Schmidt AM. N(epsilon)-(carboxymethyl)lysine adducts of proteins are ligands for receptor for advanced glycation end products that activate cell signaling pathways and modulate gene expression. J Biol Chem. 1999;274:31740–31749. doi: 10.1074/jbc.274.44.31740. [DOI] [PubMed] [Google Scholar]
- Koch M, Chitayat S, Datillo BM, Schiefner A, Diez J, Chazin W, Fritz G. Structural Basis for Ligand recognition and activation of RAGE. Structure. 2010;18:1342–1352. doi: 10.1016/j.str.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Leclerc E, Fritz G, Weibel M, Heizmann CW, Galichet A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J Biol Chem. 2007;282:31317–31331. doi: 10.1074/jbc.M703951200. [DOI] [PubMed] [Google Scholar]
- Masse JE, Keller R. AutoLink: automated sequential resonance assignment of biopolymers from NMR data by relative-hypothesis-prioritization-based simulated logic. J Magn Reson. 2005;174:133–151. doi: 10.1016/j.jmr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Yoshida T, Murata H, Harada S, Fujita N, Nakamura S, Yamamoto Y, Watanabe T, Yonekura H, Yamamoto H, et al. Solution structure of the variable-type domain of the receptor for advanced glycation end products: new insight into AGE-RAGE interaction. Biochemistry. 2008;47:12299–12311. doi: 10.1021/bi800910v. [DOI] [PubMed] [Google Scholar]
- Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- Nogi T, Sangawa T, Tabata S, Nagae M, Tamura-Kawakami K, Beppu A, Hattori M, Yasui N, Takagi J. Novel affinity tag system using structurally defined antibody-tag interaction: application to single-step protein purification. Protein Sci. 2008;17:2120–2126. doi: 10.1110/ps.038299.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostendorp T, Leclerc E, Galichet A, Koch M, Demling N, Weigle B, Heizmann CW, Kroneck PM, Fritz G. Structural and functional insights into RAGE activation by multimeric S100B. Embo J. 2007;26:3868–3878. doi: 10.1038/sj.emboj.7601805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins SJ, Wuthrich K. Ring current effects in the conformation-dependent NMR chemical shifts of aliphatic protons in the basic pancreatic trypsin inhibitor. Biochim. Biophys. Acta. 1979;576:409–423. doi: 10.1016/0005-2795(79)90416-1. [DOI] [PubMed] [Google Scholar]
- Povey JF, Howard MJ, Williamson RA, Smales CM. The effect of peptide glycation on local secondary structure. J. Struct. Biol. 2008;161:151–161. doi: 10.1016/j.jsb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Ramasamy R, Vannucci SJ, Yan SS, Herold K, Yan SF, Schmidt AM. Advanced glycation end products and RAGE: a common thread in aging, diabetes, neurodegeneration, and inflammation. Glycobiology. 2005a;15:16R–28R. doi: 10.1093/glycob/cwi053. [DOI] [PubMed] [Google Scholar]
- Ramasamy R, Yan SF, Schmidt AM. The RAGE axis and endothelial dysfunction: maladaptive roles in the diabetic vasculature and beyond. Trends Cardiovasc Med. 2005b;15:237–243. doi: 10.1016/j.tcm.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Schimmel PR, Cantor CR. Biophysical Chemistry: PartII; techniques for the Study of Biologycal Structure and Function. New York: W.H. Freeman; 1980. [Google Scholar]
- Schmidt AM, Hofmann M, Taguchi A, Yan SD, Stern DM. RAGE: a multiligand receptor contributing to the cellular response in diabetic vasculopathy and inflammation. Semin Thromb Hemost. 2000;26:485–493. doi: 10.1055/s-2000-13204. [DOI] [PubMed] [Google Scholar]
- Schmidt AM, Hori O, Cao R, Yan SD, Brett J, Wautier JL, Ogawa S, Kuwabara K, Matsumoto M, Stern D. RAGE: a novel cellular receptor for advanced glycation end products. Diabetes. 1996;45 Suppl 3:S77–S80. doi: 10.2337/diab.45.3.s77. [DOI] [PubMed] [Google Scholar]
- Schmidt AM, Hori O, Chen JX, Li JF, Crandall J, Zhang J, Cao R, Yan SD, Brett J, Stern D. Advanced glycation endproducts interacting with their endothelial receptor induce expression of vascular cell adhesion molecule-1 (VCAM-1) in cultured human endothelial cells and in mice. A potential mechanism for the accelerated vasculopathy of diabetes. J Clin Invest. 1995;96:1395–1403. doi: 10.1172/JCI118175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AM, Stern DM. Receptor for age (RAGE) is a gene within the major histocompatibility class III region: implications for host response mechanisms in homeostasis and chronic disease. Front Biosci. 2001;6:D1151–D1160. doi: 10.2741/schmidt. [DOI] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Sugaya K, Fukagawa T, Matsumoto K, Mita K, Takahashi E, Ando A, Inoko H, Ikemura T. Three genes in the human MHC class III region near the junction with the class II: gene for receptor of advanced glycosylation end products, PBX2 homeobox gene and a notch homolog, human counterpart of mouse mammary tumor gene int-3. Genomics. 1994;23:408–419. doi: 10.1006/geno.1994.1517. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ. Cell activation by glycated proteins. AGE receptors, receptor recognition factors and functional classification of AGEs. Cell Mol Biol (Noisy-le-grand) 1998;44:1013–1023. [PubMed] [Google Scholar]
- Thornalley PJ. Clinical significance of glycation. Clin Lab. 1999;45:263–273. [Google Scholar]
- Thornalley PJ, Battah S, Ahmed N, Karachalias N, Agalou S, Babaei-Jadidi R, Dawnay A. Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. Biochem J. 2003;375:581–592. doi: 10.1042/BJ20030763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente T, Gella A, Fernandez-Busquets X, Unzeta M, Durany N. Immunohistochemical analysis of human brain suggests pathological synergism of Alzheimer's disease and diabetes mellitus. Neurobiol Dis. 37:67–76. doi: 10.1016/j.nbd.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Wa C, Cerny RL, Clarke WA, Hage DS. Characterization of glycation adducts on human serum albumin by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chim Acta. 2007;385:48–60. doi: 10.1016/j.cca.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Burz DS, He W, Bronstein IB, Lednev I, Shekhtman A. Hexameric calgranulin C (S100A12) binds to the receptor for advanced glycated end products (RAGE) using symmetric hydrophobic target-binding patches. J Biol Chem. 2007;282:4218–4231. doi: 10.1074/jbc.M608888200. [DOI] [PubMed] [Google Scholar]
- Xie J, Reverdatto S, Frolov A, Hoffmann R, Burz DS, Shekhtman A. Structural basis for pattern recognition by the receptor for advanced glycation end products (RAGE) J Biol Chem. 2008;283:27255–27269. doi: 10.1074/jbc.M801622200. [DOI] [PubMed] [Google Scholar]
- Zong H, Madden A, Ward M, Mooney MH, Elliott CT, Stitt AW. Homodimerization is essential for the receptor for advanced glycation end products (RAGE)-mediated signal transduction. J Biol Chem. 285:23137–23146. doi: 10.1074/jbc.M110.133827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for Measurement of Intermolecular NOEs by Multinuclear NMR Spectroscopy: Application to a Bacteriophage λ N-Peptide/boxB RNA Complex. J Am Chem Soc. 1997;119:6711–6721. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01