Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 10.
Published in final edited form as: Structure. 2011 Aug 10;19(8):1138–1148. doi: 10.1016/j.str.2011.06.001
SUMMARY
CCL5 (RANTES) is a pro-inflammatory chemokine known to activate leukocytes through its receptor, CCR5. Although the monomeric form of CCL5 is sufficient to cause cell migration in vitro, CCL5’s propensity for aggregation is essential for migration in vivo, T cell activation and apoptosis, and HIV entry into cells. However, there is currently no structural information on CCL5 oligomers larger than the canonical CC chemokine dimer. In this study, the solution structure of a CCL5 oligomer was investigated using an integrated approach including NMR residual dipolar couplings to determine allowed relative orientations of the component monomers, SAXS to restrict overall shape, and hydroxyl radical footprinting and NMR cross saturation experiments to identify interface residues. The resulting model of the CCL5 oligomer provides a basis for explaining the disaggregating effect of E66 and E26 mutations and suggests mechanisms by which glycosaminoglycan binding may promote oligomer formation and facilitate cell migration in vivo.
INTRODUCTION
Chemokines are an important class of immuno-active signaling molecules which activate leukocytes through chemokine-family G protein-coupled receptors (GPCRs). They are widely expressed by many cells in different contexts including immune surveillance and inflammation. In all cases, the establishment of chemokine concentration gradients on endothelial layers and in the surrounding tissue provides directional cues to guide cell movement. One factor that may be vital to the establishment and maintenance of a gradient is the chemokines’ ability to oligomerize (Appay et al., 1999; Czaplewski et al., 1999). Although monomeric mutants of chemokines are capable of causing cell migration in vitro (Laurence et al., 2000; Paavola et al., 1998; Proudfoot et al., 2003; Rajarathnam et al., 1994), several studies using non-oligomerizing variants have shown that induction of migration in vivo is dependent on oligomerization (Appay et al., 1999; Campanella et al., 2006; Johnson et al., 2004; Proudfoot et al., 2003). Moreover, chemokines that are unable to oligomerize have shown therapeutic benefit in a number of animal models of inflammatory disease (Braunersreuther et al., 2010; Johnson et al., 2004), underscoring the importance of understanding oligomeric interactions.
While some chemokines can form large oligomers in isolation, oligomerization of chemokines is promoted by interaction with glycosaminoglycans (GAGs) which are sulfated polysaccharides that reside on cell surfaces and in the extracellular matrix. The importance of chemokine:GAG interactions was demonstrated experimentally when chemokine mutants competent to bind receptor, but incapable of binding GAGs, were shown to be unable to activate cell migration in vivo, presumably because GAG interactions are also important for establishing a haptotactic chemokine gradient (Johnson et al., 2004; Murooka et al., 2006; Proudfoot et al., 2003). Chemokine:GAG interactions have also been shown to be important for processes separate from migration such as T-cell activation and induction of apoptosis (Johnson et al., 2004; Murooka et al., 2006; Proudfoot et al., 2003).
Structural knowledge of chemokine oligomers alone and in complex with GAGs could clearly provide insight into the molecular mechanisms by which chemokines facilitate the recruitment and activation of leukocytes in vivo. But there is currently relatively little information on the mechanisms of chemokine oligomerization. Where models for quaternary assemblies of chemokines larger than dimers do exist, they have been deduced from crystal structures (Hoover et al., 2000; Jabeen et al., 2008; Jin et al., 2010; Lau et al., 2004; Murphy et al., 2010; Ren et al., 2010; Swaminathan et al., 2003). Most recently, Ren et al. solved the crystal structures of MIP-1α/β oligomers (Ren et al., 2010), and while SAXS data support the presence of similar structures in solution, this is one of the few cases where such experimental evidence exists. For many chemokines, there are no data on structures of oligomers in either crystals or solution.
CCL5 is a particularly aggregation-prone member of the chemokine family, and one for which no structural data on higher oligomers exist. This pro-inflammatory chemokine is secreted by both endothelial cells and activated leukocytes to attract leukocytes to sites of inflammation. In addition to migration, oligomerization of CCL5 has been shown to be important for its ability to cause generalized T cell activation through protein tyrosine kinases and for induction of apoptosis (Appay et al., 1999; Murooka et al., 2006). Furthermore, these processes were shown to involve G-protein(Gαi)-independent activation of its receptor CCR5 (Appay et al., 1999; Murooka et al., 2006) suggesting that oligomeric CCL5 activates different signaling pathways than the those induced by monomeric CCL5. Thus monomeric and oligomeric CCL5 appear to act as “functionally selective” or “biased” ligands of CCR5, terms usually reserved for different ligands of the same receptor (Keov et al.).
In addition to cell migration, CCR5 plays a crucial role in mediating HIV entry into T cells and monocytes. In this context, differences in oligomerization state also have functional consequences. While high concentrations of aggregation competent WT CCL5 promote HIV entry, low concentrations of CCL5 and mutants incapable of forming large aggregates suppress HIV infection (Czaplewski et al., 1999). Inhibition of HIV entry with low concentrations of CCL5 or mutants is thought to involve binding of monomeric CCL5 to CCR5, by physically blocking access to the receptor, and causing receptor internalization; and indeed, variants with a greater propensity to internalize have been shown to be more potent inhibitors of HIV entry (Escola et al., 2010). By contrast, high concentrations of oligomerization competent CCL5 seem to enhance HIV entry by physical crosslinking of HIV particles to target cells or by activation of kinase signaling pathways through binding to the GAG chains of the proteoglycan CD44, which in turn impacts the HIV life cycle (Roscic-Mrkic et al., 2003). Hence, understanding oligomerization tendencies is potentially important to both control of immune response and intervention in HIV infection.
Physical characterization of the oligomerization behavior of CCL5 has previously revealed that it forms large aggregates at μM concentrations in a neutral pH environment but smaller aggregates at reduced pH. The fact that large aggregates dissociate into dimers at pH 3–4, as well as the existence of mutants that limit oligomerization (Czaplewski et al, 1999), has allowed dimer characterization by both NMR and X-ray crystallography (Chung et al., 1995; Skelton et al., 1995). Structural studies have also been carried out on other CCL5 mutants and complexes with GAG disaccharides under conditions that favor dimers (Jin et al., 2010; Shaw et al., 2004; Wilken et al., 1999). But, none of the structures provide insight into the quaternary conformation of CCL5 higher order oligomers.
Utilizing wild type (WT) CCL5’s relatively high solubility at an intermediate pH (4.5), we now present CCL5’s higher oligomeric structures using solution NMR, SAXS and hydroxyl radical footprinting mass spectrometry (MS). NMR and SAXS data revealed that the size distribution of the CCL5 oligomer at pH 4.5 is primarily a tetramer. Relative orientations of the dimeric units within the tetramer were determined by NMR residual dipolar couplings (RDC) data which significantly restricts the number of possible configurations that the tetramer can adopt. Shape information from SAXS was then used to find the best translational placement of the dimers within the tetramer. The interaction interfaces between the dimers were confirmed using both NMR cross-saturation experiments and hydroxyl radical footprinting MS. Together these data provide information, not only on the assembly of the tetramer, but on the formation of linear higher order aggregates. The resulting model shows that both GAG binding sites and residues known to interact with CCR5 are exposed in a linear array, possibly facilitating movement of leukocytes along cell surfaces and aiding pathogen entry in vivo.
RESULTS
Symmetry of the WT CCL5 tetramer
The oligomerization state of WT CCL5 at a concentration of ~1mM and pH 4.5 was investigated using pulse gradient diffusion NMR, correlation times from NMR spin relaxation, dynamic light scattering, and SAXS data. NMR data clearly showed diffusional properties consistent with a tetramer. Light scattering and SAXS data indicated the presence of some higher oligomers in addition to tetramers (Supplemental figure 1 and supporting text). A two dimensional 1H-15N HSQC (heteronuclear single quantum coherence) spectrum of an 15N-labeled CCL5 sample under these conditions (Supplemental figure 2) revealed only one set of cross-peaks, one for each protonated amide in the sequence, suggesting that the oligomers formed are symmetric structures, at least when averaged over the NMR timescale.
Determination of structures for symmetric complexes can be approached with NOE based NMR experiments, but only with mixed isotopic labeling strategies and experiments that have low sensitivity (Clore and Gronenborn, 1998). Instead we opted to employ RDC measurements (Al-Hashimi et al., 2000; Prestegard et al., 2004). The resulting data are sensitive to orientations of N-H bond vectors relative to the magnetic field and provide both a test for preservation of the monomer structure as observed by X-ray crystallography (Tjandra and Bax, 1997; Tolman et al., 1995) and information on the orientation of monomers in higher order structures (Al-Hashimi et al., 2000). RDCs for WT CCL5 aligned in both neutral and positively charged polyacrylamide gel and the dimer forming E66S mutant aligned in neutral polyacrylamide gel were measured. Not surprisingly the RDCs for the E66S mutant fit very well to those back-calculated from a monomer as it appears in the dimer of the crystal structure (correlation coeff. 0.96, Q-factor 0.24). More surprisingly, both sets of RDC data collected on the WT CCL5 tetramer fit the crystal structure nearly as well (Supplemental figure 3, correlation coeff. 0.96 & 0.95, Q-factor 0.26 & 0.29 for neutral and positively charged gel RDC respectively). This indicates preservation of the dimer structure within the tetramer structure.
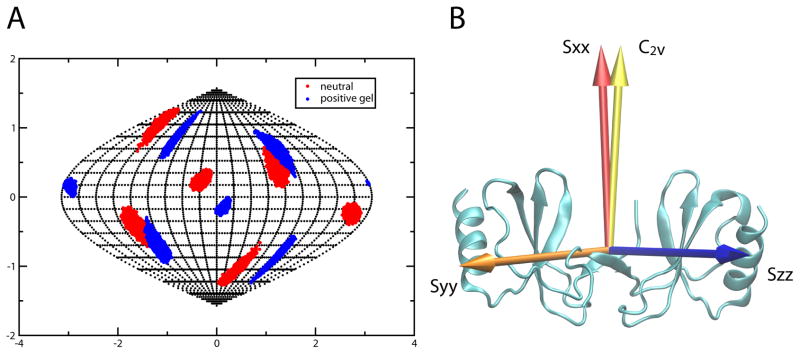
RDCs can also be used to identify an axis of rotational symmetry. For the E66S mutant one axis of the principal alignment frame determined from RDC data very closely aligned to the C2V symmetry axis observed in the crystal structure. The alignment tensor orientation and C2V symmetry axis for the WT CCL5 tetramer was determined independently by comparing alignment frame directions from data collected in neutral and positively charged gels. Figure 1A shows the directions of principle alignment frame axes for both media as viewed from the crystal structure coordinate frame. The projection plot shows that only one of the axis directions is preserved (the X-axis). As a C2V symmetry axis must be parallel to an alignment frame principal axis irrespective of alignment media (Al-Hashimi et al., 2000), this unambiguously identifies the symmetry axis of the tetramer. Figure 1B shows the orientation of the alignment frame for WT CCL5 aligned in positively charged gel. The dimer symmetry axis as seen in the crystal structure differs by only 5.5° from the X-axis of the alignment frame. The principal Sxx order parameter for WT CCL5 aligned in positively charged gel is −1.23×10−4, while the order parameters for the Y- and Z-axes are nearly equal in magnitude (Syy=−8.82×10−4, Szz=1.01×10−3) leading to an asymmetry parameter of 0.75. Comparison, of order parameters for WT CCL5 and the E66S dimer aligned in a neutral gel show similar asymmetry parameters but slightly lower order for the dimer (Sxx=0.11×10−4, Syy=6.30×10−4 and Szz=−6.41×10−4 vs. Sxx =0.70×10−4, Syy=7.40×10−4 and Szz=−8.09×10−4).
Figure 1.
A) Sauson-Flamsteed projection plot of alignment frame orientations for WT CCL5 aligned in both positively charged and neutral polyacrylamide gel. The X-axes of the two frames share a common orientation. B) Orientation of the alignment tensor relative to the CCL5 dimer. The X-axis of the principal axes of the alignment tensor is shown in red, the Y-axis is shown in blue and the Z-axis is in orange. The golden arrow indicates the orientation of the symmetry axis from the dimer crystal structure. (see also Figure S2 and S3)
The identification of the symmetry axis and preservation of the dimer structure places substantial restrictions on the structure of the tetramer. Except for a special case in which dimers stack along the symmetry axis, all allowed tetramers must have inter-dimer binding interfaces that are restricted to a band of residues that make contact through translation of one member of a properly oriented dimer pair in the YZ plane. This restriction arises because translational motion parallel to the symmetry axis would break the symmetry. The special case of a stacked tetramer is unlikely since order parameters suggest that the overall shape of the tetramer is similar to that of the dimer and the stacked version would be much more globular. Elimination of the globular structure was also confirmed using SAXS data, as described below.
Shape restrictions from SAXS data
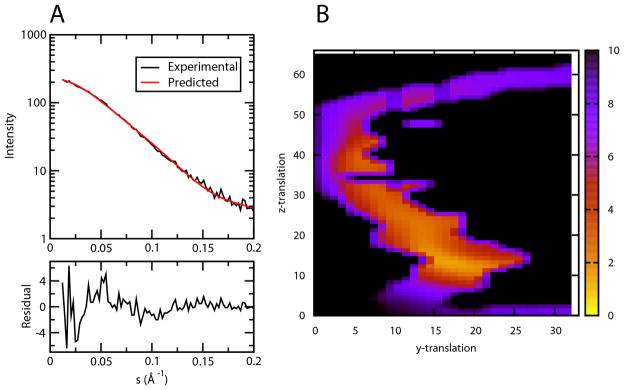
SAXS data are particularly sensitive to the spatial distribution of electron density of the dominant species in solution. Data were collected on a sample containing ~ 1.25 mM WT CCL5 at pH 4.5. The resulting scattering curve is shown in Figure 2A along with fits to models as described below. Among the parameters most relevant to distinguishing models are the radius of gyration from a Guinier plot and the maximum pair-wise distance between heavy atoms from a pair density distribution function which have values of ~31 Å and ~121 Å respectively (Supplemental figure 4). The radius of gyration is consistent with an in-plane tetramer model, and particularly, the maximum pair-wise distance is more consistent with the 122 Å extension of an in-plane hexamer than the 70 Å extension of a stacked dimer model.
Figure 2.
A) Fitting to SAXS scattering data for model from grid point 19×13, χ=1.13, 40% of the protein is assumed in be in the tetramer form. B) Contour map of the SAXS fitting χ values of the models generated on the grid. Colors ranging from blue to yellow represent high to low χ values. (see also Figure S4)
Modeling using a symmetry-restricted grid search, SAXS & residue pairing score
Given the restrictions on the symmetry and shape of the tetramer, a simple grid search was used to generate all possible models satisfying the symmetry constraints, using a procedure similar to that described by Wang et al. (Wang et al., 2008). However, additional degrees of freedom were added to allow small symmetry preserving rotations around the Y- & Z-axes (15 and 5 degrees, respectively) to take into account errors in determining the symmetry axis. The models were subsequently screened for agreement with the SAXS scattering profile (Figure 2B) and a favorable binding surface using a residue-pairing score (Supplemental figure 5) (Moont et al., 1999). During the SAXS profile fitting, the presence of a tetramer-hexamer equilibrium was treated through the use of the program OLIGOMER (Konarev et al., 2003). The hexamer model was produced by adding an additional dimer unit to the tetramer, duplicating the initial dimer-dimer interface, and then adjusting the tetramer-hexamer ratio to find the best fit to the SAXS data. Monomers and dimers were not included in the simulations; the dimer dissociation constant for the wild type at pH 3.8 has been determined to be 18 μM (Duma et al., 2007; Skelton et al., 1995). This suggests that only a small percentage of monomers are present at concentrations used in the SAXS experiments. Figure 2B shows the contour map of χ values of the SAXS fitting for models produced at each grid point. To improve selectivity, a score combining both SAXS χ values and a residue pair score was created to select models (see Figure 3A & Methods). One group of models centered around the grid point 19×13 gave the best combined score. The best fit of model 19×13 to the experimental SAXS scattering curve was obtained by assuming 40% of the protein mass existed as tetramer and the rest as hexamer. Figure 2A shows the comparison of the theoretically calculated scattering curve of the hexamer/tetramer mixture of model 19×13 with the experimental scattering curve.
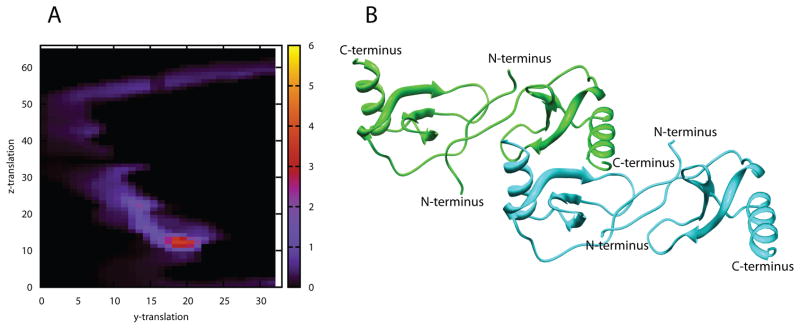
Figure 3.
A) Contour maps of the combined residue pair and SAXS scores for each model on the grid. B) Model from grid point 19×13, which is representative of the models from the group with the best (orange region) combined score. (see also Figure S5)
Figure 3B shows the model from grid point 19×13, which is representative of the models from the cluster surrounding this point. The tetramer model is based on an interface formed by contacts between residues 25 to 30 in the second β-strand from one monomer of dimer A and several residues of the C-terminal helix from one monomer of dimer B. A symmetric set of interactions is contributed by the β-strand of dimer B and the C-terminal helix of dimer A. Models generated from other grid points in the cluster differ from one another only in small displacements along the Y- and Z-axes.
Validating the binding interface with NMR cross saturation data
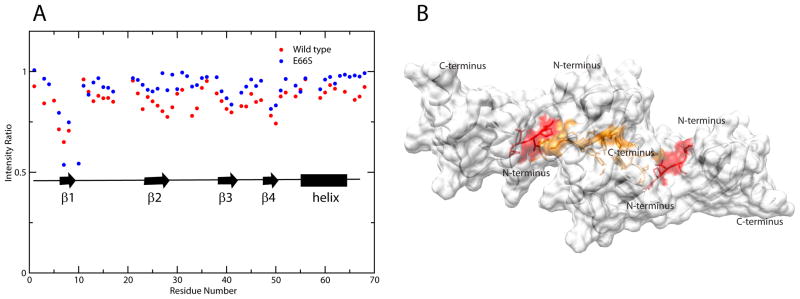
To validate the model found in the grid search, the inter-dimer interface was probed using solution NMR cross saturation (Takahashi et al., 2000). In this method, 15N, 2H-labeled CCL5 was mixed with unlabeled CCL5 in a 1:3 ratio, and mixed dimers and tetramers were allowed to form. By applying RF pulses to the methyl/methylene region of the proton spectrum, sites in the unlabeled molecules were saturated and spin diffusion transferred this saturation across the interface to the nearest 1H−15N pairs of labeled molecules. The ratios of HSQC cross-peak intensities, with and without saturation, provide a measure of the proximity of various 1H−15N pairs to the interface. Both dimer and tetramer interfaces will be seen, so it is important to compare experiments for the E66S mutant and WT to distinguish monomer-monomer from dimer-dimer interfaces. Figure 4A shows a plot of HSQC saturated/unsaturated signal intensity ratios for WT and E66S CCL5. Although the two sets of data are not quantitatively comparable due to differences in rotational correlation times and resulting differences in spin diffusion efficiencies, they still serve as a good qualitative guide for identifying the dimer-dimer interface in the tetramer. For WT CCL5, the intensities of N-terminal residues 6 to 8 (red in Figure 4B), 10, 15 to 17, 23, 26 to 29, 33–34, and 49–50 are highly reduced. Additional intensity changes were seen for residues 40 to 43 and 64 to 68. Intensity decreases for N-terminal residue signals are clearly due to interactions at the monomer-monomer interface in the dimer units since the same residues are perturbed in spectra of the E66S mutant. Residues 26 to 29, 33 and 34, and 64 to 68 were perturbed in the WT data only (orange area in Figure 4B); they are in the dimer-dimer interface of the tetramer model and clearly support this model. Perturbation of residues 40 to 43 and 48 to 51 are seen in both proteins and can be explained by the fact that these residues are in the vicinity of the monomer-monomer interface and may be indirectly saturated by their proximity to the interfacial residues. The weak saturation of residues 15 to 17, as well as 23, seemed perplexing at first; however, the N-terminus of CCL5 is extremely flexible as shown by NMR dynamics measurements (Supplemental figure 1B) and the conformation of its end is undefined in both crystal and solution structures of CCL5. In view of these observations, it is conceivable that the N-terminal residues may transiently interact with residues 15 to 17 and 23 on the dimeric partner, leading to saturation. On the whole, the binding interface defined by the cross saturation experiment is in agreement with the model found through the SAXS/RDC grid search procedure.
Figure 4.
A) Plot of residue specific cross saturation-induced amide proton signal intensity changes for WT and E66S CCL5. B) Surface plot of the CCL5 tetramer model with residues identified as being specifically perturbed in the wild type (residues 26 to 29, 33, 34, 66 and 67) colored in orange and the dimer interface (residues 6 to 10) colored in red.
Validating the binding interface through hydroxyl radical footprinting MS
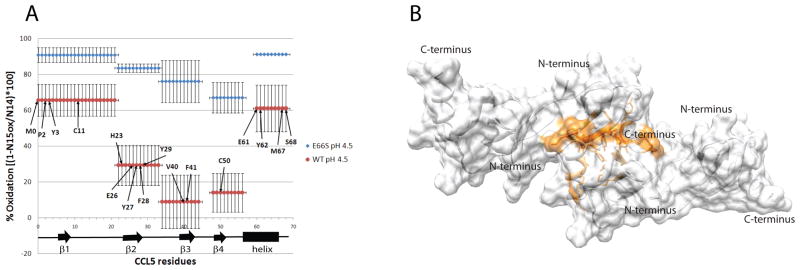
The binding interface of the CCL5 tetramer was also validated through hydroxyl radical footprinting MS. UV radiation-induced hydrogen peroxide radiolysis was used to produce hydroxyl radicals that rapidly oxidize exposed protein side chains, and the degree of oxidation was interpreted as an indicator of relative solvent exposure. By comparing the solvent exposure of the E66S mutant, which is a mixture of monomer and dimer at the concentrations studied in this experiment (20 μM and pH 4.5), with WT under the same conditions, regions that are protected in the dimer-dimer interface of the tetramer can be identified by a decrease in the rate of oxidation. In this experimental set up, the radical was produced and consumed on a sub-microsecond time scale to ensure that modifications stemming from non-native side chain exposures, i.e. as a result of conformational changes caused by oxidation of the protein, were kept to a minimum (Gau et al., 2009). For quantitation, the amount of oxidation of each peptide within CCL5 was determined by measuring the amount of unmodified peptide compared to an isotopically labeled control, then MS/MS was used to determine the major residues oxidized for each peptide, providing amino acid level resolution. Figure 5A compares the peptides and residues protected in WT CCL5 to those protected in the E66S mutant. H23, E26, Y27, F28, Y29, V40, F41, and C50 were found to be highly protected from oxidation in the WT structures compared to the E66S dimer, while oxidation of M1, P2, Y3, and C11 on the N-terminal peptide and E61, Y62, M67, and S68 on the C-terminal peptide are only slightly more protected. M0, P2, Y3, C11, H23, V40 and C50 appear to be protected in the monomer-monomer interface of the dimer, and their increases in protection may reflect a depletion of monomer concentration or some additional stabilization of the dimer on oligomerization. Figure 5B shows the locations of the remaining protected residues on the surface of the CCL5 tetramer. It is evident that the binding surface identified by protection of these residues from hydroxyl radical footprinting agrees very well with that of the cross saturation experiment.
Figure 5.
A) Plot of residue specific hydroxyl radical modification percentage for WT and E66S CCL5. The degree of modification is analyzed at the peptide level, and the major sites of oxidation are identified at the residue level for each peptide. Residues identified as being protected from modification are indicated. B) Surface plot of the CCL5 tetramer with residues identified as being in the tetrameric interface (residues 26 to 29, 41, 61, 62, 67, 68,) colored orange. (see also Figure S6)
Formation of higher order oligomers
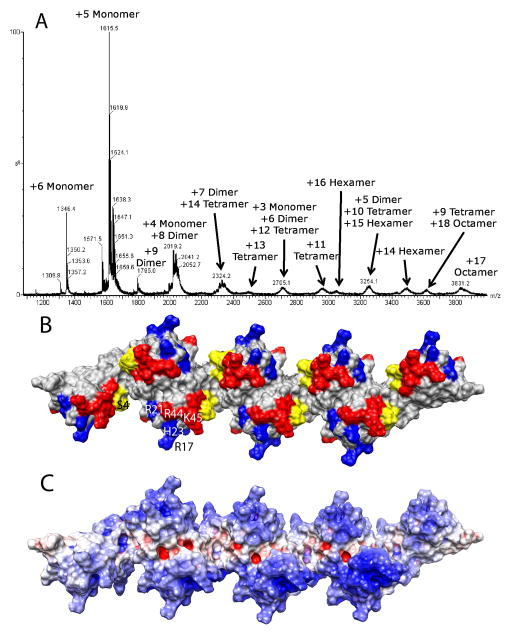
The nature of the interface in the tetramer structures shown in Figures 3–5 is one that leads naturally to formation of higher order oligomers. Use of residues from only one monomer of each dimer leaves those residues in the second monomer open to interactions that propagate the structure to linear polymers of even order (hexamers, octamers, etc). In addition to SAXS data indicating a substantial amount of hexamer at the somewhat higher concentrations used in those experiments, native spray MS performed on WT CCL5 at pH 4 indicated the presence of higher oligomers (Figure 6A). Even though these data were acquired at lower injection concentrations (~10μM), the loss of solvent from electrospray droplets is known to promote and allow detection of oligomers that normally form at higher concentrations. In addition to large amounts of monomer and dimer observed, significant amounts of tetramer, hexamer and octamer were also detected, but not trimer, pentamer or heptamer. These results indicate that the higher order oligomers are a result of a concatenation of a dimer substructure, rather than oligomerization of the monomer structure (which would lead to odd-numbered oligomers), or oligomerization of a unique tetramer structure (which would not allow for the formation of a hexamer). Further support for progressive oligomerization using the same dimer-dimer interface is seen in hydroxyl radical footprinting data on a WT CCL5 sample at pH 7 where a higher level of oligomerization is observed (Supplemental figure 6). An overall increase in protection is seen, possibly due to a decrease in monomer content and in the percentage of exposed dimer ends with increased oligomer size. More importantly, the highest percentage increase in protection was observed for the 22–33 peptide that contains several of the residues suggested to comprise the dimer-dimer interface of the tetramer.
Figure 6.
A) Native spray mass spectrum of WT CCL5 (10 μM) at pH 4.5. Even-numbered oligomers from dimer to octamer are observed, indicating that the oligomer is built from a concatenation of dimer substructures. B) Surface plot of the CCL5 octamer model with residues perturbed by GAGs (residues 44–48, 55, 56) shown in red and residues known to contact CCR5 at pH 6 (residues 16, 17, 21, 23) shown in blue. The N-terminus of CCL5, which is both perturbed by GAGs and known to bind to CCR5 in CCL5 monomer is colored yellow. C) Electrostatic potential plot of the octamer surface showing large patches of basic regions (blue) throughout the protein.
Figure 6B shows a model of an octamer constructed by joining four dimers using the same interface contacts identified in the tetramer. The type of interaction employed in generating this model would normally propagate indefinitely. The fact that we observe restriction to fairly small oligomers is likely correlated with the high positive charge of CCL5 (pI of 9). The increasing net charge with increase in oligomer size would serve to decrease the association constant for addition of each new dimer. Figure 6C shows the electrostatic potential surface plot of the CCL5 octamer. It is evident from the extensive positive patches on the complex that large electrostatic repulsions can be generated, particularly at pH 4.5 where additional protonation of some carboxylate groups can occur. The situation may, however, be quite different in the presence of negatively charged GAGs and at more physiological pH, as suggested from analytical ultracentrifugation studies (Czaplewski et al., 1999) and native spray MS of WT CCL5 at pH 7, which revealed a very broad, unresolved peak consistent with the presence of very large, highly charged and heterogeneous oligomers (data not shown).
DISCUSSION
The studies presented here suggest a compelling oligomerization model for the pro-inflammatory chemokine CCL5. The model was generated by integrating complementary data from multiple techniques, including existing crystal structures of dimers, solution NMR data, SAXS data and MS hydroxyl radical footprinting. The results show that CCL5 is capable of oligomerizing without significantly changing its dimeric structure or the orientation of a C2V symmetry axis. SAXS data are inconsistent with the formation of more globular tetramer structures, such as those observed for MCP-1 and IP-10 (Lau et al., 2004; Swaminathan et al., 2003). Instead, the resulting model is elongated, providing a mechanism for propagating interactions among dimer units into long linear polymeric chains containing an even number of monomer units.
Molecular basis of the dimer-dimer interaction
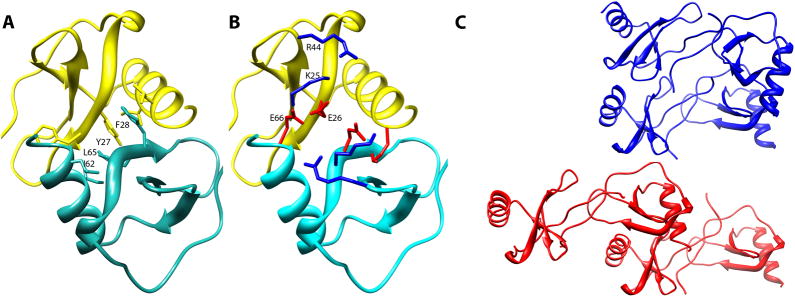
The interaction interface identified through a combination of hydroxyl radical footprinting and NMR cross-saturation data is primarily composed of contacts between residues 25 to 30 in the second β-strand and residues at the C-terminal helix of monomer A of one dimer and similar residues in the neighboring dimer. The contact surface is approximately 860 Å2 and is stabilized by a mixture of hydrophobic and electrostatic interactions at the interface. In particular, Y27 and F28 are situated in the middle of the interface with Y27s on separate dimer units forming aromatic ring stacking interactions. F28 provides a further hydrophobic interaction surface by making cross unit contacts with and L65 and I62 on the C-terminus (Figure 7A). The interface also appears to be stabilized by an intermolecular salt bridge between K25 on one dimer unit and E26 from the neighboring unit, with both centered at the interface. E66 is also positioned close to K25 as well as R44 in the 40s loop and likely exerts additional stabilization forces on the interaction through the formation of salt bridges (Figure 7B). The involvement of E66 and E26 in the interface provides a rationalization for the reduced oligomerization tendency of mutants in which these residues are changed to serine or alanine (Czaplewski et al., 1999).
Figure 7.
A) Details of the inter-dimer hydrophobic interactions in model 19×13. The hydrophobic interface is formed by Y27, F28, I62 and L65. B) Electrostatic interactions at the dimer-dimer interface. K25, E26, E66 & R44 can form pairs of electrostatic bonds. C) Ribbon representations of the proposed CCL5 tetramer (red) and the MIP1α tetramer (blue). A single dimer unit from each tetramer is arranged in identical orientation.
Comparison of the CCL5 tetramer to other tetramer structures
While there are no structures of oligomers larger than a dimer for CCL5, a number of higher order oligomeric structures have been observed in crystal structures of other chemokines. Given the conservation of monomer topologies across all four families of chemokines, and the tendency of most CC chemokines to form similar “CC-like dimers” and CXC chemokines to form “CXC-like dimers”, there is a surprising number of different higher order oligomerization topologies represented in structural studies of chemokines. Small sequence variations seem to have large consequences on the stable form of higher order oligomers with structures varying from more globular forms for tetramers (Lau et al., 2004; Swaminathan et al., 2003) to more extended forms where chains as long as decamers are observed (Murphy et al., 2010; Ren et al., 2010). Our more extended CCL5 model has a topology most closely resembling oligomers seen in the recently published crystal structure of a polymeric form of MIP-1α/β by Ren et al. (Ren et al., 2010). The relative orientation of the dimers in the MIP-1 polymers is very similar to that of CCL5 and some of the same residues are involved in forming the tetrameric interface in these structures. However, the dimer units are translated in our model making it somewhat more extended (Figure 7C). This translation produced a much better fit with the experimental SAXS scattering profile than a model based directly on the MIP-1α/β structure. The structural differences between CCL5 and MIP-1 oligomers appear to arise primarily from sequence variation. Although they share 42% sequence identity, CCL5 is far more basic than MIP-1α: the pI for MIP-1α/β is 4.8 whereas the pI for CCL5 is 9.2. The more basic nature of CCL5 may prevent it from oligomerizing into a more compact MIP-1α-like structure which would bring positively charged residues in close proximity, creating unfavorable interactions. Furthermore, K25, which is not present in MIP-1α, appears to be crucial to the formation of the interface in CCL5, acting as a bridge for the formation of inter-dimer electrostatic interactions with both E26 and E66. Without K25, the only favorable electrostatic interactions may be between the GAG-binding motif and E26/E66, as seen in the MIP-1α oligomer structures. The presence of GAGs would then compete with the formation of higher oligomers. The difference in these electrostatic properties and residue specific interactions may explain the experimental observation that GAGs can reduce the oligomerization of WT MIP-1α while leaving the oligomerization state of its D27A mutant, which forms an unstable tetramer, untouched (Ren et al., 2010). These same GAG-binding residues play only a partial role in the formation of the CCL5 tetramer and are more exposed in the D27A mutant of MIP1α.
Even greater sequence variations between CCL5 and chemokines such as MCP-1 may explain why CCL5 does not form a MCP-1 type globular tetramer. Despite having a greater number of charged residues, MCP-1 has neither the K25/E26 pair nor the 40s BBXB motifs found in CCL5. The lack of these residues may preclude the formation of CCL5/MIP-1 -type linear oligomers, since these motifs play an important role in the formation of the interface. This leaves MCP-1 to utilize the “alternate CXC dimer interface” at its N-terminus to form the globular tetramer. It also should be noted that formation of CCL5 tetramers similar to MCP-1 would result in the burial of critical GAG binding residues in the 40s loop, which would be both thermodynamically destabilizing and would preclude interactions with GAGs.
Predicted interactions with glycosaminoglycans
Given the importance of chemokine oligomerization and GAG binding in leukocyte migration, it is appropriate to consider how interactions with GAGs may occur in the context of our current model, and how they affect the oligomerization process. Prominent among the residues believed to be involved in GAG binding are the BBXB motifs constituting residues K44, K45, N46, R47, and to a lesser extent, K55 and K56, all of which are highlighted in red in the octamer model of WT CCL5 (Figure 6B). Notably, all of the residues are solvent accessible and their linear arrangement on the surface of the CCL5 oligomer forms sites on which GAG chains could bind. This arrangement therefore offers an explanation for GAG-promoted chemokine oligomerization (Hoogewerf et al., 1997; Vivès et al., 2002). In the absence of GAGs, the electrostatic repulsive force would keep the size of the CCL5 oligomer small. However, binding of sulfated GAGs would neutralize repulsive forces between CCL5 dimers increasing their aggregation propensity. This may account for the ability of even low molecular weight GAGs to cause aggregation (Vivès et al., 2002; Yu et al., 2005). Perhaps equally important, the ability of GAG polymers to bridge the binding sites on multiple chemokine subunits, could have a stabilizing effect on the oligomers through avidity, somewhat akin to molecular Velcro. Similarly, chemokine oligomers enhance the affinity for GAGs by providing multiple binding epitopes. Along these lines, heparin is hypothesized to stabilize an MCP-1 tetramer by bridging all four subunits whereas MCP-1 forms primarily dimers in the absence of GAGs. Furthermore, the affinity of the monomeric form of MCP-1 for GAG is reduced relative to the WT protein (Lau et al., 2004).
Simultaneous interaction with chemokine receptors and glycosaminoglycans
It is also useful to consider how the formation of chemokine oligomers and their interactions with GAGs fits into the overall process of leukocyte extravasation. The consensus mechanism for this process stipulates that chemokine-induced firm adhesion of leukocytes to the endothelium occurs after weak selectin-mediated adhesion (Alon et al., 2003; Lutters et al., 2004; Miles et al., 2008). If true, oligomerized chemokines immobilized on the surface of the endothelium via GAGs may be the first form of chemokine exposed to receptors on leukocytes. Indeed, Weber and coworkers demonstrated that WT CCL5, but not aggregation-deficient variants, could induce CCR1-mediated leukocyte arrest, suggesting a role of chemokine oligomers in integrin activation (Baltus et al., 2003). This observation further suggests that oligomeric CCL5 bound to its receptor induces a different set of signals than the monomeric form as may be required at different stages of the process: leukocyte arrest, migration and activation (Appay et al., 1999; Baltus et al., 2003; Salanga and Handel, 2011), and thus define the different oligomeric forms as biased ligands. Some of the residues of CCL5 important for interacting with CCR5 have been identified by chemical shift perturbation from titrations with a peptide derived from the CCR5 N-terminus. At low pH (3.8), the peptide has a stronger interaction with the CCL5 monomer than the dimer and residues normally forming the monomer-monomer interface are among the most strongly perturbed (Duma et al., 2007). This is consistent with the monomer being involved in the activation of CCR5 at low concentration. However, at pH 6 the difference in monomer and dimer behavior decreases, consistent with the fact that oligomers can also interact at high concentration, and the most strongly perturbed residues of the N-terminus shift to residues 16, 17 and 23. In Figure 6B these residues are colored in blue and are shown to be fully accessible for receptor interaction.
Although never experimentally demonstrated, this line of thinking also lends itself to the concept that chemokines immobilized on surfaces can simultaneously interact with receptors and GAGs, and our model suggests mechanisms by which dual engagement could occur. First the GAG-binding residues and the CCR5 interacting residues occupy separate regions on the surface of CCL5 (Figure 6B), which could permit simultaneous interactions. Secondly, it is possible that GAGs could interact with some blocks of the polymer while receptors simultaneously interact with other blocks. However, the most interesting mechanism suggested by the model is that the linear arrangement of binding epitopes, offering sequential equivalent sites for CCR5 interaction, may promote diffusion of leukocytes along the CCL5 polymer. One dimensional diffusion offers well known advantages over diffusion in two and three dimensions and is exploited in many other cellular contexts (Gorman and Greene, 2008; Vale et al., 1989). Thus, it is reasonable to consider that facilitated diffusion on GAG chains could be operative in cell migration.
EXPERIMENTAL PROCEDURES
Expression of WT & E66S CCL5
Two versions of mature CCL5 were expressed in E. coli using a pET23a vector with conventional 15NH4Cl-containing M9 media. They were purified according to Czaplewski et al. (Czaplewski et al., 1999). Briefly, inclusion bodies of CCL5 were solubilized in 6M Gn-HCl and purified on a Superdex 75 column. Purified protein was then subjected to fast dilution refolding with refolding buffer consisting of 100mM Tris, pH 8.0, 100 μM reduced glutathione and 10 μM oxidized dimeric glutathione. The refolded protein was then dialyzed against 0.1% TFA and purified further using strong anion exchange chromatography.
NMR spectroscopy
NMR samples of CCL5 contained 1 to 2 mM of protein in 50 mM acetate buffer at pH 4.4. RDCs were measured on a 1 mM CCL5 sample aligned in stretched 5% positively charged (Cierpicki and Bushweller, 2004) or neutral polyacrylamide gels using the two-stage gel tube as described in Liu et al. (Liu and Prestegard, 2010). RDC values were measured using a J-modulated 1H,15N-HSQC experiment (Liu and Prestegard, 2010). Samples for cross-saturation measurements were conducted on a sample consisting of 0.75 mM 15N,2H-labeled protein mixed with 2.25 mM unlabeled protein in 50% D2O, 50 mM acetate, pH 4.4, buffer. Cross saturation experiments were carried out in a manner similar to that published by Takahashi et al. (Takahashi et al., 2000). Protons were either saturated at 1 ppm or −23 ppm as a control.
SAXS measurements
SAXS measurements were carried out as described by Wang et al. (Wang et al., 2011). Specifically, samples containing 1 to 1.25 mM of WT CCL5 at pH 4.5 were irradiated for one hour with a 1.54 Å X-ray source. The scattering pattern was collected using a Bruker Nanostar U X-ray system and the 2D scattering patterns of both background and sample were reduced to 1D scattering curves using Bruker’s SAXS software. The scattering profile was determined as the difference between samples with and without protein. Values for maximum pair distances were extracted using the program GNOM (Svergun, 1992).
Model screening using grid search, SAXS &, residue pairing score
The grid search was conducted in a manner similar to Wang et al. (Wang et al., 2008). Specifically, the crystal structure of the CCL5 dimer (PDB accession code 1U4L) was placed in the alignment tensor principal axes frame such that the presumed symmetry axis became the X-axis of the PDB frame. Another dimer of CCL5, obtained by rotating the first dimer by 180 º around the X-axis, was then translated over a grid on a plane perpendicular to the X-axis. Theoretical SAXS scattering curves of each tetramer/hexamer model obtained by placing the second dimer at a different grid point was calculated using CRYSOL (Svergun et al., 1995) and a hydration shell contrast value of 0.07 e/Å3. The curves were then used in OLIGOMER (Konarev et al., 2003) to find the best fit to a tetramer/hexamer mixture. The interface of each model was evaluated using a residue pairing score (Moont et al., 1999) and all models were evaluated using a combined score, which is obtained by scaling the residue pairing score to be in the same numeric range as the SAXS fitting χ values for models with a χ value of 2.50 or less.
Hydroxyl radical footprinting MS
Samples of 15N-labeled CCL5 (E66S mutant or wild type) at 20 μM in acetate buffer (pH 4.5) containing 50 μM H2O2 with 20 μM glutamine to limit radical half-life was flowed through the beam path of a KrF laser at 248 nm, and pulsed so that each segment of sample was irradiated with a single ~20 ns UV pulse with a small buffer region between irradiated segments to help account for sample diffusion and laminar flow effects. The oxidation was immediately quenched using methionine amide (0.5 μg/μl) and catalase (0.5 μg/μl). Prior to trypsin digestion, unoxidized and unlabeled mutant or WT CCL5 (20 μM) was added to the corresponding irradiated samples to serve as an internal standard for quantitation. After trypsin digestion FT-MS was used to derive unmodified, unlabeled peptide peaks to unmodified, 15N labeled peptide peak ratios for each tryptic peptide, and the fractional oxidation was calculated from the reduction of ratios compared to an internal standard. Tandem mass spectrometry was used in conjunction with LC-MS to identify sites of oxidation, with sites of oxidation determined by manual annotation of MS/MS spectra.
Supplementary Material
01
HIGHLIGHTS.
- Wild type CCL5 oligomerizes to even order structures in a pH dependent manner.
- A tetramer structure was determined by integrating NMR, SAXS and MS data.
- Higher order oligomers are formed by dimers ordered in a staggered line.
- The model exposes GAG- and CCR5-binding residues, for simultaneous interaction.
Acknowledgments
We thank Bruker AXS for use of their NANOSTAR system and Brian Jones for collecting and evaluating the bioSAXS data on WT CCL5. We thank Dr. Hsiau-Wei Lee for assistance with RDC collection. We also acknowledge the National Center for Research Resources (a part of the NIH) for financial support of the Resource for Integrated Glycotechnology and the CCL5 project (P41-RR005351) and the National Institute of General Medical Sciences K99 program for support of X.W. (K99GM088483) ), and the National Institute of Allergy and Infectious Disease for support of T.M.H. (RO1AI37113 and AI037113-13S1). The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
SUPPLEMENTAL INFORMATION AND ACCESSION CODES
Protein Data Bank: Coordinates have been deposited under accession number 2L9H.
Note: Supplementary information is available on the Structure website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Hashimi HM, Bolon PJ, Prestegard JH. Molecular symmetry as an aid to geometry determination in ligand protein complexes. J Magn Reson. 2000;142:153–158. doi: 10.1006/jmre.1999.1937. [DOI] [PubMed] [Google Scholar]
- Alon R, Grabovsky V, Feigelson S. Chemokine induction of integrin adhesiveness on rolling and arrested leukocytes local signaling events or global stepwise activation? Microcirculation. 2003;10:297–311. doi: 10.1038/sj.mn.7800195. [DOI] [PubMed] [Google Scholar]
- Appay V, Brown A, Cribbes S, Randle E, Czaplewski LG. Aggregation of RANTES is responsible for its inflammatory properties - Characterization of nonaggregating, noninflammatory RANTES mutants. Journal of Biological Chemistry. 1999;274:27505–27512. doi: 10.1074/jbc.274.39.27505. [DOI] [PubMed] [Google Scholar]
- Baltus T, Weber KS, Johnson Z, Proudfoot AE, Weber C. Oligomerization of RANTES is required for CCR1-mediated arrest but not CCR5-mediated transmigration of leukocytes on inflamed endothelium. Blood. 2003;102:1985–1988. doi: 10.1182/blood-2003-04-1175. [DOI] [PubMed] [Google Scholar]
- Braunersreuther V, Pellieux C, Pelli G, Burger F, Steffens S, Montessuit C, Weber C, Proudfoot A, Mach F, Arnaud C. Chemokine CCL5/RANTES inhibition reduces myocardial reperfusion injury in atherosclerotic mice. J Mol Cell Cardiol. 2010;48:789–798. doi: 10.1016/j.yjmcc.2009.07.029. [DOI] [PubMed] [Google Scholar]
- Campanella GS, Grimm J, Manice LA, Colvin RA, Medoff BD, Wojtkiewicz GR, Weissleder R, Luster AD. Oligomerization of CXCL10 is necessary for endothelial cell presentation and in vivo activity. J Immunol. 2006;177:6991–6998. doi: 10.4049/jimmunol.177.10.6991. [DOI] [PubMed] [Google Scholar]
- Chung CW, Cooke RM, Proudfoot AEI, Wells TNC. The 3-Dimensional Solution Structure of Rantes. Biochemistry. 1995;34:9307–9314. doi: 10.1021/bi00029a005. [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Bushweller JH. Charged gels as orienting media for measurement of residual dipolar couplings in soluble and integral membrane proteins. Journal of the American Chemical Society. 2004;126:16259–16266. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- Czaplewski LG, McKeating J, Craven CJ, Higgins LD, Appay V, Brown A, Dudgeon T, Howard LA, Meyers T, Owen J, et al. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1 alpha, MIP-1 beta, and RANTES - Characterization of active disaggregated chemokine variants. Journal of Biological Chemistry. 1999;274:16077–16084. doi: 10.1074/jbc.274.23.16077. [DOI] [PubMed] [Google Scholar]
- Duma L, Haussinger D, Rogowski M, Lusso P, Grzesiek S. Recognition of RANTES by extracellular parts of the CCR5 receptor. J Mol Biol. 2007;365:1063–1075. doi: 10.1016/j.jmb.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Escola JM, Kuenzi G, Gaertner H, Foti M, Hartley O. CC chemokine receptor 5 (CCR5) desensitization: cycling receptors accumulate in the trans-Golgi network. J Biol Chem. 2010;285:41772–41780. doi: 10.1074/jbc.M110.153460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gau BC, Sharp JS, Rempel DL, Gross ML. Fast Photochemical Oxidation of Protein Footprints Faster than Protein Unfolding. Analytical Chemistry. 2009;81:6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nature Structural & Molecular Biology. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- Hoogewerf AJ, Kuschert GSV, Proudfoot AEI, Borlat F, ClarkLewis I, Power CA, Wells TNC. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- Hoover DM, Mizoue LS, Handel TM, Lubkowski J. The crystal structure of the chemokine domain of fractalkine shows a novel quaternary arrangement. J Biol Chem. 2000;275:23187–23193. doi: 10.1074/jbc.M002584200. [DOI] [PubMed] [Google Scholar]
- Jabeen T, Leonard P, Jamaluddin H, Acharya KR. Structure of mouse IP-10, a chemokine. Acta Crystallographica Section D-Biological Crystallography. 2008;64:611–619. doi: 10.1107/S0907444908007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kagiampakis I, Li P, Liwang PJ. Structural and functional studies of the potent anti-HIV chemokine variant P2-RANTES. Proteins. 2010;78:295–308. doi: 10.1002/prot.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Z, Kosco-Vilbois MH, Herren S, Cirillo R, Muzio V, Zaratin P, Carbonatto M, Mack M, Smailbegovic A, Rose M, et al. Interference with heparin binding and oligomerization creates a novel anti-inflammatory strategy targeting the chemokine system. Journal of Immunology. 2004;173:5776–5785. doi: 10.4049/jimmunol.173.9.5776. [DOI] [PubMed] [Google Scholar]
- Keov P, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors: A pharmacological perspective. Neuropharmacology. 60:24–35. doi: 10.1016/j.neuropharm.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- Lau EK, Paavola CD, Johnson Z, Gaudry JP, Geretti E, Borlat F, Kungl AJ, Proudfoot AE, Handel TM. Identification of the glycosaminoglycan binding site of the CC chemokine, MCP-1 - Implications for structure and function in vivo. Journal of Biological Chemistry. 2004;279:22294–22305. doi: 10.1074/jbc.M311224200. [DOI] [PubMed] [Google Scholar]
- Laurence JS, Blanpain C, Burgner JW, Parmentier M, LiWang PJ. CC chemokine MIP-1 beta can function as a monomer and depends on Phe13 for receptor binding. Biochemistry. 2000;39:3401–3409. doi: 10.1021/bi9923196. [DOI] [PubMed] [Google Scholar]
- Liu YZ, Prestegard JH. A device for the measurement of residual chemical shift anisotropy and residual dipolar coupling in soluble and membrane-associated proteins. Journal of Biomolecular Nmr. 2010;47:249–258. doi: 10.1007/s10858-010-9427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutters BCH, Leeuwenburgh MA, Appeldoorn CCM, Molenaar TJM, Van Berkel TJC, Biessen EAL. Blocking endothelial adhesion molecules: a potential therapeutic strategy to combat atherogenesis. Curr Opin Lipidology. 2004;15:545–552. doi: 10.1097/00041433-200410000-00008. [DOI] [PubMed] [Google Scholar]
- Miles FL, Pruitt FL, van Golen KL, Cooper CR. Stepping out of the flow: capillary extravasation in cancer metastasis. Clin Exp Metastasis. 2008;25:305–324. doi: 10.1007/s10585-007-9098-2. [DOI] [PubMed] [Google Scholar]
- Moont G, Gabb HA, Sternberg MJ. Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins. 1999;35:364–373. [PubMed] [Google Scholar]
- Murooka TT, Wong MM, Rahbar R, Majchrzak-Kita B, Proudfoot AEI, Fish EN. CCL5-CCR5-mediated apoptosis in T cells - Requirement for glycosaminoglycan binding and CCL5 aggregation. Journal of Biological Chemistry. 2006;281:25184–25194. doi: 10.1074/jbc.M603912200. [DOI] [PubMed] [Google Scholar]
- Murphy JW, Yuan H, Kong Y, Xiong Y, Lolis EJ. Heterologous quaternary structure of CXCL12 and its relationship to the CC chemokine family. Proteins. 2010;78:1331–1337. doi: 10.1002/prot.22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola CD, Hemmerich S, Grunberger D, Polsky I, Bloom A, Freedman R, Mulkins M, Bhakta S, McCarley D, Wiesent L, et al. Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J Biol Chem. 1998;273:33157–33165. doi: 10.1074/jbc.273.50.33157. [DOI] [PubMed] [Google Scholar]
- Prestegard JH, Bougault CM, Kishore AI. Residual dipolar couplings in structure determination of biomolecules. Chem Rev. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- Proudfoot AEI, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TNC, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarathnam K, Sykes BD, Kay CM, Dewald B, Geiser T, Baggiolini M, Clark-Lewis I. Neutrophil activation by monomeric interleukin-8. Science. 1994;264:90–92. doi: 10.1126/science.8140420. [DOI] [PubMed] [Google Scholar]
- Ren M, Guo Q, Guo L, Lenz M, Qian F, Koenen RR, Xu H, Schilling AB, Weber C, Ye RD, et al. Polymerization of MIP-1 chemokine (CCL3 and CCL4) and clearance of MIP-1 by insulin-degrading enzyme. EMBO J. 2010 doi: 10.1038/emboj.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscic-Mrkic B, Fischer M, Leemann C, Manrique A, Gordon CJ, Moore JP, Proudfoot AE, Trkola A. RANTES (CCL5) uses the proteoglycan CD44 as an auxiliary receptor to mediate cellular activation signals and HIV-1 enhancement. Blood. 2003;102:1169–1177. doi: 10.1182/blood-2003-02-0488. [DOI] [PubMed] [Google Scholar]
- Salanga CL, Handel TM. Chemokine oligomerization and interactions with receptors and glycosaminoglycans: The role of structural dynamics in function. Exp Cell Res. 2011 doi: 10.1016/j.yexcr.2011.01.004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JP, Johnson Z, Borlat F, Zwahlen C, Kungl A, Roulin K, Harrenga A, Wells TNC, Proudfoot AEI. The X-ray structure of RANTES: Heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure. 2004;12:2081–2093. doi: 10.1016/j.str.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Skelton NJ, Aspiras F, Schall TJ. Solution Structure of Rantes, a C-C Chemokine. Journal of Cellular Biochemistry. 1995:37–37. doi: 10.1021/bi00016a004. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- Svergun DI. DETERMINATION OF THE REGULARIZATION PARAMETER IN INDIRECT-TRANSFORM METHODS USING PERCEPTUAL CRITERIA. Journal of Applied Crystallography. 1992;25:495–503. [Google Scholar]
- Swaminathan GJ, Holloway DE, Colvin RA, Campanella GK, Papageorgiou AC, Luster AD, Acharya KR. Crystal structures of oligomeric forms of the IP-10/CXCL10 chemokine. Structure. 2003;11:521–532. doi: 10.1016/s0969-2126(03)00070-4. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Nakanishi T, Kami K, Arata Y, Shimada I. A novel NMR method for determining the interfaces of large protein-protein complexes. Nat Struct Biol. 2000;7:220–223. doi: 10.1038/73331. [DOI] [PubMed] [Google Scholar]
- Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins: information for structure determination in solution. Proc Natl Acad Sci U S A. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Soll DR, Gibbons IR. ONE-DIMENSIONAL DIFFUSION OF MICROTUBULES BOUND TO FLAGELLAR DYNEIN. Cell. 1989;59:915–925. doi: 10.1016/0092-8674(89)90614-4. [DOI] [PubMed] [Google Scholar]
- Vivès RR, Sadir R, Imberty A, Rencurosi A, Lortat-Jacob H. A Kinetics and Modeling Study of RANTES(9–68) Binding to Heparin Reveals a Mechanism of Cooperative Oligomerization. Biochemistry. 2002;41:14779–14789. doi: 10.1021/bi026459i. [DOI] [PubMed] [Google Scholar]
- Wang X, Bansal S, Jiang M, Prestegard JH. RDC-assisted modeling of symmetric protein homo-oligomers. Protein Sci. 2008;17:899–907. doi: 10.1110/ps.073395108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee HW, Liu Y, Prestegard JH. Structural NMR of protein oligomers using hybrid methods. J Struct Biol. 2011;173:515–529. doi: 10.1016/j.jsb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilken J, Hoover D, Thompson DA, Barlow PN, McSparron H, Picard L, Wlodawer A, Lubkowski J, Kent SBH. Total chemical synthesis and high-resolution crystal structure of the potent anti-HIV protein AOP-RANTES. Chemistry & Biology. 1999;6:43–51. doi: 10.1016/S1074-5521(99)80019-2. [DOI] [PubMed] [Google Scholar]
- Yu Y, Sweeney MD, Saad OM, Crown SE, Hsu AR, Handel TM, Leary JA. Chemokine-glycosaminoglycan binding: specificity for CCR2 ligand binding to highly sulfated oligosaccharides using FTICR mass spectrometry. J Biol Chem. 2005;280:32200–32208. doi: 10.1074/jbc.M505738200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01