Direct Detection of Staphylococcus Osteoarticular Infections by Use of Xpert MRSA/SA SSTI Real-Time PCR (original) (raw)
Abstract
We evaluated the Xpert MRSA/SA SSTI real-time PCR assay (Cepheid, Sunnyvale, CA) directly on perioperative bone and joint samples. The sensitivity and specificity for detection of methicillin-susceptible Staphylococcus aureus (MSSA), methicillin-resistant Staphylococcus aureus (MRSA), and methicillin-resistant coagulase-negative Staphylococcus were, respectively, 100% and 98.3%, 100% and 100%, and 100% and 95.3%. The median total test turnaround time was 72 min for PCR versus 79 h for culture. Using these rapid results, appropriate antibiotic treatment could be rapidly initiated.
INTRODUCTION
The reported incidence of septic arthritis (SA) varies from 2 to 5 cases/100,000 individuals annually in the general population to 40 to 68 cases/100,000 individuals annually among patients with joint prostheses (26, 35). While septic arthritis is often clinically demonstrative (fever, erythema, swelling, decreased range of motion, etc.), diagnosis of prosthetic joint infections (PJI) is often delayed because of its diverse symptomatology (20). Staphylococcus aureus is the most prominent pathogen in the vast majority of cases of suppurative acute arthritis in adults and in children >2 years of age. In contrast, coagulase-negative staphylococci are frequently isolated in chronic arthritis, such as prosthetic joint infections (30, 34). In all clinical isolates, methicillin resistance occurs frequently (in about 50% to 65% of the strains), as recently demonstrated by Frazee et al. (16).
Several studies have evaluated the role of PCR in the diagnosis of osteoarticular infections (11, 12, 15, 28). However, most of the PCR methods used have limitations, such as complex technical requirements, extended hands-on time and test turnaround time, and poor specificity and sensitivity, which represent barriers to routine use. Furthermore, to date, none of these assays has been able to detect antibiotic resistance at the same time.
Recently, the Xpert MRSA/SA SSTI real-time PCR assay on GeneXpert has become commercially available and has been FDA cleared and CE (Communauté Européenne) marked for the detection of methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) in skin and soft tissue infections due to the simultaneous detection of three targets (spa, SCC_mec_, and mecA). The random-access GeneXpert performs extraction, amplification, and detection of the targets in a single-use cartridge so that results are available in about 56 min after a sample is received (45).
The aim of our study was to evaluate the Xpert MRSA/SA SSTI real-time PCR assay (Cepheid, Sunnyvale, CA) directly on perioperative samples obtained during surgery.
MATERIALS AND METHODS
Patients and specimens.
A total of 135 samples from 105 patients were prospectively analyzed from March 2009 to July 2010, representing prosthetic joint infections (46), spondylodiscitis (3), acute septic arthritis (15), and noninfected controls (33).
For all patients, medical records were evaluated for the following: demographic characteristics; clinical, radiographical, biological, histopathological, and microbiological data; type of surgical management; and antimicrobial therapy.
The study was approved by the Institutional Review Board of the Clinique de L'Union.
Study definition.
(i) Prosthetic joint infections. Patients were considered to have PJI when at least one of the following criteria was present: (a) visible purulence of a preoperative aspirate or intraoperative periprosthetic tissue (as determined by the surgeon), (b) a sinus tract communicating with the prosthesis, (c) acute inflammation in intraoperative permanent prosthetic tissue sections by histopathology (as determined by the pathologist), (d) an increased synovial fluid leukocyte count with ≥1,700 leukocytes/μl and/or ≥65% polymorphonuclear neutrophils in the joint fluid, or (e) microbial growth in preoperative joint aspirate or intraoperative periprosthetic tissues (1, 38, 39, 41, 47).
(ii) Acute septic arthritis. Patients with at least one of the following criteria were considered to be suffering from septic arthritis: (a) a short (1- to 2-week) history of pain, swelling, and restricted movement in the affected joint; (b) an increased synovial fluid leukocyte count with ≥1,700 leukocytes/μl; (c) ≥65% polymorphonuclear neutrophils in the joint fluid; or (d) microbial growth in the synovial fluid (35).
(iii) Spondylodiscitis. Spondylodiscitis was considered as a diagnosis when at least one of the following criteria was fulfilled: (a) spine pain with elevation of inflammatory serum markers with or without fever, (b) evocative computed tomography (CT) or magnetic resonance imaging (MRI) results, or (c) microbial growth in blood cultures and/or discovertebral biopsy specimens (19).
(iv) Negative controls. Patients who underwent revision or resection of their bone and joint prostheses (hip, knee, and shoulder) between March 2009 and July 2010 and did not show any septic symptomatology or bacterial growth in the perioperative samples were included as controls.
Processing of samples.
Synovial fluids were aspirated preoperatively by the operating surgeon and transferred into a sterile vial (DS Laboratoires, France). Tissue specimens were collected in the same sterile vials.
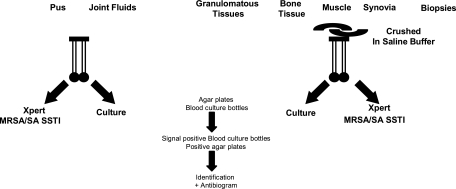
Culture and Xpert MRSA/SA SSTI assays were performed simultaneously for every patient, as shown in Fig. 1.
Fig. 1.
Processing of samples. A double swab was directly immersed in the liquid samples, while solid samples were previously crushed in saline buffer. One of the swabs was immediately submitted to Xpert MRSA/SA SSTI assay, while the other was plated. Liquid samples were also inoculated in blood culture bottles. Any single colony was identified, and an antibiogram was performed.
In PJI, according to expert recommendations, 3 to 5 samples, including synovial fluid and periprosthetic tissue biopsy specimens, were collected (36, 37, 43). For 15 patients, only 2 of these samples were analyzed, according to the protocol mentioned above.
Histopathology was performed on all samples. Cultures were analyzed by workers separate from those performing the PCR assay, and the results of either assay were not available to those performing the other test method.
Microbiological cultures.
Tissue specimens (bone, muscles, fascia, etc.) were scratched, dilacerated, and then homogenized by vortexing in 1 ml sterile saline solution for 1 min, while synovial fluids were vortexed and inoculated directly. Both samples were divided into two aliquots.
A Copan liquid Stuart swab (part no. 900-0370; Cepheid, Sunnyvale, CA) was immersed for 5 s in the first aliquot, and then the swab was broken into the MRSA/SA SSTI cartridge according to the manufacturer's recommendations (see below). It has been suggested that such a swab picks up around 0.1 ml of sample (13).
The second aliquot was dedicated to microbial cultures. In the case of synovial fluid, the leukocyte count and proteins were previously determined. Ten microliters of the aliquots was plated onto aerobic and anaerobic sheep blood agar plates (bioMérieux, Marcy l'Etoile, France), which were incubated for 14 days at 35°C ± 2°C in 5% CO2 and under anaerobiosis, respectively. Residual volumes were inoculated into BacTalert aerobic and anaerobic bottles and incubated for 14 days in a BactTalert system (bioMérieux, Marcy l'Etoile, France) according to French and American expert guidelines (36, 37, 43). All signal-positive bottles were seeded on agar plates and incubated in an adequate atmosphere.
Each unique colony of isolated microorganisms was identified using standard microbiological techniques (Vitek 2 Compact and API Galeries; bioMérieux, Marcy l'Etoile, France), and its antimicrobial susceptibility was tested. Methicillin susceptibility was determined using P581 cards (Vitek 2 Compact; bioMérieux, Marcy l'Etoile, France), and discrepant results were confirmed by testing a cefoxitin disk (30 μg) according to Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM) recommendations (6, 7), as well as PBP2a (penicillin-binding protein 2a) detection (Slidex MRSA detection; bioMérieux, Marcy l'Etoile, France).
Multiplex PCR assay.
Real-time PCR was performed using the MRSA/SA SSTI assay on the fully automated GeneXpert system (Cepheid, Sunnyvale, CA). The extraction, amplification, and detection steps take place in different chambers of a self-contained, single-use cartridge containing all the reagents necessary for the detection of the bacterial targets. The primers and probes detect sequences within the staphylococcal protein A (spa) gene, the gene for methicillin resistance (mecA), and the staphylococcal cassette chromosome (SCC_mec_) inserted into the S. aureus chromosomal attB insertion site, as well as an internal-control sample processing control (SPC) (Bacillus globigii). The MRSA/SA SSTI PCR was performed according to the manufacturer's instructions. Briefly, samples were adsorbed onto a Copan liquid Stuart's swab (Cepheid part no. 900-370) from the container received in the laboratory, transferred into the sample reagent vial, and treated according to the manufacturer's recommendations. The cartridge was then placed into the GeneXpert instrument, and the test was performed according to the MRSA/SA SSTI program. The overall process is complete in 56 min and can be performed in a random-access manner.
Several amplification profiles could be obtained. When spa, mecA, and SCC_mec_ were amplified, MRSA presence in the sample was established, whereas when spa and SCC_mecA_ were amplified, MSSA was present in the sample. Finally, when mecA was the only amplified target, the presence of a methicillin-resistant coagulase-negative staphylococcus (MRCoNS) in the sample was suspected (this result is not mentioned in the product insert). In all cases, the amplification of SPC (B. globigii internal control) was checked. All these molecular results were compared to standard culture results.
Hands-on time was measured according to the mapping process method. Each step was noted and evaluated with a chronometer. The hands-on time for culture was longer than expected because of the processing of solid samples into suspensions before inoculation on media.
Data analysis.
Standard culture was considered the gold standard. The sensitivity, specificity, and positive (PPV) and negative (NPV) predictive values were determined. The median test hands-on time and turnaround time were also evaluated. Statistical analysis was performed (Excel 2007 and Statview II software), and P values of ≤0.05 were considered significant.
RESULTS
Characteristics of the patients.
The median patient ages were comparable in all the studied groups, ranging from 57 years (infected-patient group) to 64 years in the control group. Males were predominant in the infected-patient group (70.1%) with respect to the control group (64.7%) (Table 1).
Table 1.
Characteristics of patients
| Characteristic | Infected patients | Controls |
|---|---|---|
| Demographics (no.) | 72 | 33 |
| Type of infection | ||
| PJI | 55 | |
| Septic arthritis | 14 | |
| Spondylodiscitis | 3 | |
| Prosthetic joint revision | 33 | |
| Age [median (range)] (yr) | 57 (8–83) | 64 (19–89) |
| Sex (no.) | ||
| Male | 44 | 22 |
| Female | 18 | 12 |
| Location of arthritis [no. (%)] | ||
| Elbow | 3 (4.7) | 0 |
| Knee | 16 (25.3) | 11 (33.3) |
| Hip | 22 (34.9) | 20 (60.6) |
| Ankle | 6 (9.4) | 6 (18.2) |
| Shoulder | 10 (15.8) | 1 (3.0) |
| Wrist | 2 (3.1) | 0 |
| Interphalangeal joint | 1 (1.5) | 0 |
| Vertebral disc | 3 (4.7) | 0 |
| Origin of samples (no.) | ||
| Synovial fluid | 35 | 14 |
| Bone | 14 | 12 |
| Adjacent tissues | 11 | 8 |
| Vertebral disc | 3 | 0 |
| Biological data | ||
| WBCa at admission (no./mm3) | 10,500 | 6,500 |
| Fibrinogen at admission (g/liter) | 4.29 | 3.09 |
| CRP at admission (mg/liter) | 56 | 6 |
| Previous antibiotherapy | ||
| No. of patients | 14 | 0 |
| Median duration (days) | 8 |
In order to be exhaustive, all kinds of osteoarticular perioperative samples from various types of joints were included in the study. Knee, hip, ankle, and shoulder, however, were most often represented. The types of samples were also diversified, as synovial fluid, bone biopsy specimens, periprosthetic tissues, and discovertebral biopsy specimens were analyzed.
Biological data showed an increase of leukocytes in septic arthritis and spondylodiscitis (12,100 and 11,800/mm3, respectively), while fibrinogen was supranormal exclusively in SA (6.03 g/liter). Furthermore, C-reactive protein (CRP) was moderately elevated in PJI and spondylodiscitis (SP) (respectively, 26 and 14 mg/liter) but dramatically increased in SA (128 mg/liter).
Finally, 14 of the 135 (10.4%) patients had previously received antimicrobial therapy, with a median duration of 8 days (range, 1 to 60 days).
Microbiology.
Table 2 summarizes the bacteria isolated from infected patients (prosthetic joint infections, septic arthritis, and spondylodiscitis). A single causative organism was found in 57 cases (95%) and a polymicrobial infection in 3 cases (5%). Most monobacterial infections were staphylococcal infections. Methicillin resistance was detected in 27 of 53 (50.9%) staphylococcal infections, mostly in coagulase-negative staphylococcal episodes (74.2% of the strains). One infection was caused by Escherichia coli and 6 by Propionibacterium acnes, mostly in chronic shoulder PJI.
Table 2.
Comparison of microorganisms detected by standard microbiological cultures versus Xpert MRSA/SA assay
| Bacteriumb | No. | No. of isolates identifieda | |
|---|---|---|---|
| Both techniques | Culture only | PCR only | |
| Staphylococcus infections | 57 | ||
| Staphylococcus aureus | 18 | 0 | 0 |
| MRSA | 2 | ||
| MSSA | 16 | ||
| Staphylococcus epidermidis | 19 | 7 | 4 |
| MRSE | 19 + 4c | ||
| MSSE | 7 | ||
| Staphylococcus warneri | 2 | 0 | 0 |
| MRSW | 2 | ||
| MSSW | 0 | ||
| Staphylococcus capitis | 2 | 2 | 0 |
| MRSC | 0 | ||
| MSSC | 2 | ||
| Staphylococcus simulans | 2 | 0 | 0 |
| MRSS | 2 | ||
| MSSS | 0 | ||
| Other bacteria | 7 | ||
| E. coli | 0 | 1 | 0 |
| P. acnes | 0 | 6 | 0 |
| Negative controls | 91 | 2 | 0 |
None of the samples of patients included as controls showed positivity either in standard culture or with PCR (data not shown).
Comparison of culture and PCR for staphylococcal infections.
As shown in Table 2, among 64 cases of osteoarticular infections, 57 were due to staphylococci. Gram stains were positive in only 21 cases (33%). The causative organisms were detected by both Xpert MRSA/SA SSTI PCR and standard culture in 44 (77%) cases. With regard to the 13 discrepancies, the causative organism was detected by culture only in 9 cases (15.7%) (exclusively methicillin-susceptible coagulase-negative staphylococci that are not targeted by the probes). On the other hand, PCR (mecA) was the only positive assay in 4 (7%) cases.
Finally, all controls were concordant and negative for the two techniques, except for two patients who were determined to be false positive for PCR, as no infection history or bacterial growth could be observed. The patients who tested negative by Xpert MRSA/SA SSTI all had positive internal-control amplification, indicating that there were no PCR inhibitors present in the samples.
Table 3 shows a comparison of the MRSA/SA SSTI assay and standard culture for the detection of MSSA, MRSA, and MRCoNS. MRSA and MSSA presence could be directly obtained from the MRSA/SA SSTI assay, while MRCoNS was extrapolated by the amplification of mecA only. The sensitivity and specificity for detection of MSSA, MRSA, and methicillin-resistant coagulase-negative Staphylococcus were, respectively, 100% and 98.3%, 100% and 100%, and 100% and 95.3%. Statistical analysis did not show any difference between the MRSA/SA SSTI assay and standard culture and indicated high correlation between the methods.
Table 3.
Comparison of Xpert MRSA/SA SSTI assay versus standard culture for staphylococcal osteoarticular infections based on detection of MSSA, MRSA, and MRCoNS in samples
| Assay result | No. of samples with results | Assay performance [% (95% CIa) | ||||
|---|---|---|---|---|---|---|
| Positive | Negative | Sensitivity | Specificity | PPV | NPV | |
| MRSA/SA SSTI assay | ||||||
| MSSA | ||||||
| Positive | 16 | 2 | 100 (97–100) | 98.3 (92.4–100) | 88.9 (81.1–96.7) | 100 (97–100) |
| Negative | 0 | 119 | ||||
| MRSA | ||||||
| Positive | 2 | 0 | 100 (82–100) | 100 (97–100) | 100 (82–100) | 100 (97–100) |
| Negative | 0 | 133 | ||||
| MRCoNS | ||||||
| Positive | 23 | 4 | 100 (97–100) | 95.3 (90,6–100) | 85.2 (78–92) | 100 (96–100) |
| Negative | 0 | 81 | ||||
| Standard culture | ||||||
| Positive | 57 | 0 | ||||
| Negative | 4 | 74 |
We also decided to compare the hands-on times and the turnaround times of the two techniques. The median measured hands-on times for standard culture and the Xpert MRSA/SA SSTI assay were, respectively, 25 min and 2 min, while test turnaround times from admission to notification of results were, respectively, 79 h and 72 min.
DISCUSSION
Antimicrobial therapy in bone and joint infections has to be initiated as early as possible in order to prevent a negative outcome and involves a prolonged course. Empirical treatment covering most pathogens causing bacterial arthritis will normally be prescribed after obtaining a sample for laboratory studies. The choice of drugs currently depends on the Gram stain and also on risk factors (prosthesis, recent surgery, corticosteroids, etc.). Since more community-acquired infections in the United States and elsewhere are now caused by methicillin-resistant staphylococci (including S. aureus and CoNS), recommendations for treatment of infections in such patients include 1 g vancomycin every 12 h (17).
In order to achieve a rapid diagnosis of osteoarticular infections, the use of PCR amplification has been proposed for broad-range detection of bacteria in synovial fluids. However, nearly all PCR assays involve laborious and time-consuming analytical processing, making them impractical for routine clinical use (1, 24, 33, 46). A further limitation of these assays is the use of broad-range 16S rRNA gene PCR, which can detect previously unknown organisms but has lower sensitivity and specificity than specific PCR, requires subsequent bacterial identification, and fails to detect antimicrobial resistance (5, 10, 11, 12, 22, 28).
In this context, we evaluated Xpert MRSA/SA SSTI for the detection of MSSA, MRSA, and MRCoNS directly in perioperative osteoarticular samples. The goal of this study was first to validate the method for samples that were not initially included in the FDA clearance process (synovial fluids, bone and muscle biopsy specimens, and discovertebral biopsy specimens).
Gram stains were positive in only 33% of the PCR-positive/culture-positive samples, confirming that Gram stain is a poor tool for rapid clinical management of the patient. These results are consistent with the literature, as the sensitivity of direct bacterial visual examination in bone and joint samples varies from 29 to 50% (8, 14, 27).
Among all positive samples, only 3 were polymicrobial (two S. aureus strains, two CoNS strains, and a combination of Staphylococcus epidermidis and P. acnes). Fifty-seven infections were attributable to staphylococci and 7 to other bacterial species, such as E. coli and P. acnes. The latter group were obviously missed by multiplex PCR due to the lack of specific primers for these species. However, 44 (77%) were detected by both culture and PCR. This is of particular interest, as the most common specific or broad-range (16S rRNA gene) PCR methods applied to synovial fluids or periprosthetic tissues generally display a sensitivity of only 50% (10, 18, 23, 28, 42). Nine isolates were detected only by culture. These culture-positive strains were exclusively methicillin-susceptible CoNS that could not be detected, as no specific primers are included in the PCR assay.
There were four positive Xpert MRSA/SA SSTI PCR results in which microbiological culture remained negative. The only positive target was mecA. These discrepant results were obtained in two patients suffering from chronic PJI and in two other patients for whom a methicillin-resistant S. epidermidis infection had been previously diagnosed and who had been treated with antimicrobial therapy in the last 8 days before sampling. It is actually difficult to determine whether these results were false positive or whether they reflected the presence of nonviable bacteria, as PCR-positive/culture-negative results can be consistent with a number of circumstances. First, it is known that prior antibiotic therapy can lead to a negative culture. Furthermore, traditional tissue cultures might be unable to recover bacteria captured in biofilms, as well as bacteria lysed during specimen transportation or because of release of locally delivered antibiotics at the time of prosthesis removal (3, 29, 32, 39). Surgeons have been advised to discontinue antimicrobial therapy 2 to 4 weeks prior to scheduled surgical intervention if possible in order to improve the sensitivity of tissue cultures. However, some inpatients were already given antibiotics that could not be stopped at the moment of sampling. Another issue that could explain the discrepancies is the persistence of DNA from dead bacteria in samples, as it has been proven that bacterial DNA can still be detected up to 43 days after successful treatment (1, 31, 40).
When considering exclusively the staphylococcal targets detected by the two techniques (MSSA, MRSA, and, by extrapolation, mecA amplification, which could correspond to MRCoNS), the statistical performance of Xpert MRSA/SA SSTI PCR was calculated with respect to the gold standard microbiological culture. Statistical analysis of the performance did not show any difference between Xpert MRSA/SA SSTI PCR and culture performance, but rather, high correlation (P < 0.001).
Rapidity is crucial in the diagnosis of bone and joint infections. The 72-min complete turnaround time for the Xpert MRSA/SA SSTI PCR is consistent with the fact that GeneXpert is a fully automated, random-access, flexible system. This is a very short time to result, in contrast to most real-time PCR assays, which need available and well-trained operators and, consequently, batch testing. Batch PCR is rapid on paper but presents one major issue. Indeed, several studies have demonstrated that as samples are awaited in order to run the assay, results are generally available in a minimum of one working day (compared to 2 to 4 working days for culture) (2, 9, 21, 25).
These preliminary results have to be confirmed with more samples coming from distinct geographical areas (44). However, we can conclude that the Xpert MRSA/SA SSTI assay is a novel, rapid, and accurate method for the determination of many staphylococci in perioperative samples that can dramatically improve the diagnosis and the clinical management of patients suffering from bone and joint infections, as it can detect the presence of the most common species, as well as resistance to methicillin.
We suggest that the specific primers should be modified in order to include other common organisms causing osteoarticular infections, such as methicillin-susceptible coagulase-negative staphylococci, Kingella kingae, and P. acnes.
ACKNOWLEDGMENTS
We thank Cepheid for providing test kits for this study.
Cepheid had no role in the study design, data collection, data analysis, or data interpretation.
Footnotes
▿
Published ahead of print on 12 October 2011.
REFERENCES
- 1.Achermann Y., Vogt M., Leunig M., Wüst J., Trampuz A. 2010. Improved diagnosis of periprosthetic joint infection by multiplex PCR of sonication fluids from removed implants. J. Clin. Microbiol. 48:1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldeyab M. A., et al. 2009. Can the use of a rapid polymerase chain reaction screening method decrease the incidence of nosocomial methicillin-resistant Staphylococcus aureus? J. Hosp. Infect. 71:22–28 [DOI] [PubMed] [Google Scholar]
- 3.Alonzo T. A., Pepe M. S. 1999. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat. Med. 18:2897–3003 [DOI] [PubMed] [Google Scholar]
- 4.Berbari E. F., et al. 2007. Culture-negative prosthetic joint infections. Clin. Infect. Dis. 45:1113–1119 [DOI] [PubMed] [Google Scholar]
- 5.Bergin P. F., et al. 2010. Detection of periprosthetic infections with use of ribosomal RNA-based polymerase chain reaction. J. Bone Joint Surg. Am. 92:654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comité de l'Antibiogramme de la Société Française de Microbiologie. January 2009. Recommandations 2009. Société Française de Microbiologie, Paris, France: http://www.sfm-microbiologie.org/UserFiles/file/CASFM/casfm_2009-1.pdf [Google Scholar]
- 7.Comité de l'Antibiogramme de la Société Française de Microbiologie. January 2010. Recommandations 2010. Société Française de Microbiologie, Paris, France: http://nosobase.chu-lyon.fr/recommandations/sfm/2010_antibiotiques_casfm.pdf [Google Scholar]
- 8.Cuchacovich R., Quinet S., Santos A. M. 2003. Applications of polymerase chain reaction in rheumatology. Rheum. Dis. Clin. North Am. 29:1–20 [DOI] [PubMed] [Google Scholar]
- 9.Cunningham R., et al. 2007. Effect on MRSA transmission of rapid PCR testing of patients admitted to critical care. J. Hosp. Infect. 65:24–28 [DOI] [PubMed] [Google Scholar]
- 10.De Man F., et al. 2009. Broad-range PCR in selected episodes of prosthetic joint infection. Infection 37:292–294 [DOI] [PubMed] [Google Scholar]
- 11.Dempsey K. E., et al. 2007. Identification of bacteria on the surface of clinically infected and non-infected hip joints removed during revision arthroplasties by rRNA gene sequencing and microbiological culture. Arthritis Res. Ther. 9:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dora C., Altwegg M., Gerber C., Böttger E. C., Zbinden R. 2008. Evaluation of conventional microbiological techniques and molecular genetic techniques for diagnosis in patients with implanted orthopedic devices. J. Clin. Microbiol. 46:824–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake C., Barenfanger J., Lawhorn J., Verhulst S. 2005. Comparison of easy-flow Copan Liquid Stuart's and Starplex swab transport systems for recovery of fastidious aerobic bacteria. J. Clin. Microbiol. 43:1301–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faraj A. A., Omonbude O. D., Godwin P. 2002. Gram staining in the diagnosis of acute septic arthritis. Acta Orthop. Belg. 68:388–391 [PubMed] [Google Scholar]
- 15.Fihman V., et al. 2007. Improved diagnosis specificity in bone and joint infections using molecular techniques. J. Infect. 55:510–517 [DOI] [PubMed] [Google Scholar]
- 16.Frazee B. W., Fee C., Lambert L. 2009. How common is MRSA in septic arthritis? Ann. Emerg. Med. 54:695–700 [DOI] [PubMed] [Google Scholar]
- 17.Fridkin S. K., et al. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436–1444 [DOI] [PubMed] [Google Scholar]
- 18.Gallo J., et al. 2008. Culture and analysis of joint fluid in diagnosis of prosthetic joint infection. New Microbiol. 31:97–104 [PubMed] [Google Scholar]
- 19.Gouliouris T., Aliyu S. H., Brown N. M. 2010. Spondylodiscitis: update on diagnosis and management. J. Antimicrob. Chemother. 65(Suppl. 3):11–24 [DOI] [PubMed] [Google Scholar]
- 20.Gupta M. N., Sturrock R. D., Field M. 2003. Prospective comparative study of patients with culture proven and high suspicion of adult onset septic arthritis. Ann. Rheum. Dis. 62:327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harbarth S., et al. 2006. Evaluation of rapid screening and pre-emptive contact isolation for detecting and controlling methicillin-resistant Staphylococcus aureus in critical care: an interventional cohort study. Crit. Care. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hombach M., Pfyffer G. E., Roos M., Lucke K. 2010. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in specimens from various body sites: performance characteristics of the BD GeneOhm MRSA assay, the Xpert MRSA assay and broth-enriched culture in an area with a low prevalence of MRSA infections. J. Clin. Microbiol. 48:3882–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilharreborde B., et al. 2009. New real-time PCR-based method for Kingella kingae DNA detection: amplification of samples collected from 89 children with acute arthritis. J. Clin. Microbiol. 47:1837–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jalava J., Skurnik M., Tovainen A., Tovainen P., Eerola E. 2001. Bacterial PCR in the diagnosis of joint infection. Ann. Rheum. Dis. 60:287–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeyaratnam D., et al. 2008. Impact of screening tests on acquisition of methicillin-resistant Staphylococcus aureus: cluster randomized cross-over trial. BMJ 336:927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaandorp C. J. E., van Schaardenburg D., Krijnen P., Habbema J. D., van de Laar M. A. 1995. Risk factors for septic arthritis in patients with joint disease. Arthritis Rheum. 38:1819–1825 [DOI] [PubMed] [Google Scholar]
- 27.Oethinger M., Warner D. K., Schindler S. A., Kobayashi H., Bauer T. W. 2011. Diagnosing periprosthetic infection: false-positive intraoperative Gram stains. Clin. Orthop. Relat. Res. 469:954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panousis K., Grigoris P., Butcher I., Rana B., Reilly J. H., Hamblen D. L. 2005. Poor predictive value of broad-range PCR for the detection of arthroplasty infection in 92 cases. Acta Orthop. 76:341–346 [PubMed] [Google Scholar]
- 29.Patel R., Osmon D. R., Hanssen A. D. 2005. The diagnosis of prosthetic joint infection: current techniques and emerging technologies. Clin. Orthop. Relat. Res. 437:55–58 [DOI] [PubMed] [Google Scholar]
- 30.Pioro M. H., Mandell B. F. 1997. Septic arthritis. Rheum. Dis. Clin. North Am. 23:239–258 [DOI] [PubMed] [Google Scholar]
- 31.Piper K. E., et al. 2009. Microbiological diagnosis of prosthetic shoulder infection using implant sonication. J. Clin. Microbiol. 47:1878–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powles J. W., Spencer R. F., Lovering A. M. 1998. Gentamycin release from old cement during revision hip arthroplasty. J. Bone Joint Surg. Br. 80:607–610 [PubMed] [Google Scholar]
- 33.Rosey A.-L., et al. 2007. Development of broad-range 16S rDNA real-time PCR for the diagnosis of septic arthritis in children. J. Microbiol. Methods 68:88–93 [DOI] [PubMed] [Google Scholar]
- 34.Smith J. W. 1990. Infectious arthritis. Infect. Dis. Clin. North Am. 4:523–538 [PubMed] [Google Scholar]
- 35.Smith J. W., Chalupa P., Shabaz Hasan M. 2006. Infectious arthritis: clinical features, laboratory findings and treatment. Clin. Microbiol. Infect. 12:309–314 [DOI] [PubMed] [Google Scholar]
- 36.Société Française de Microbiologie. 2010. Diagnostic microbiologique des infections osseuses et articulaires, p. 165–170. In Société Française de Microbiologie (ed.), REMIC. Société Française de Microbiologie, Paris, France [Google Scholar]
- 37.Société Française de Pathologie Infectieuse de Langue Française. 2009. Recommendations for bone and joint prosthetic device infections in clinical practice (prosthesis, implants, osteosynthesis), p. 1–61. Société Française de Pathologie Infectieuse de Langue Française, Paris, France: http://www.infectiologie.com/site/consensus_recos.php [DOI] [PubMed] [Google Scholar]
- 38.Trampuz A., et al. 2004. Synovial leukocyte count and differential for the diagnosis of prosthetic knee infection. Am. J. Med. 117:556–562 [DOI] [PubMed] [Google Scholar]
- 39.Trampuz A., Piper K., Hanssen A. 2006. Sonication of explanted prosthetic components in bags for diagnosis of prosthetic joint infection is associated with risk contamination. J. Clin. Microbiol. 44:628–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trampuz A., et al. 2007. Sonication of removed hip and knee prostheses for the diagnosis of infection. N. Engl. J. Med. 357:654–663 [DOI] [PubMed] [Google Scholar]
- 41.Trampuz A., Zimmerli W. 2008. Diagnosis and treatment of implant-associated septic arthritis and osteomyelitis. Curr. Infect. Dis. Rep. 10:394–403 [DOI] [PubMed] [Google Scholar]
- 42.Vandercam B., et al. 2008. Amplification-based DNA analysis in the diagnosis of prosthetic joint infection. J. Mol. Diagn. 10:537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Versalovic J. (ed.) 2011. Manual of clinical microbiology, 10th ed American Society for Microbiology, Washington, DC [Google Scholar]
- 44.Wolk D. M., et al. 2009. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J. Clin. Microbiol. 47:758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolk D. M., et al. 2009. Rapid detection of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in wound specimens and blood cultures: multicenter preclinical evaluation of the Cepheid Xpert MRSA/SA skin and soft tissue and blood culture assays. J. Clin. Microbiol. 47:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S., et al. 2008. Rapid PCR-based diagnosis of septic arthritis by early Gram-type classification and pathogen identification. J. Clin. Microbiol. 46:1386–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmerli W., Trampuz A., Ochsner P. E. 2004. Prosthetic-joint infections. N. Engl. J. Med. 351:1645–1654 [DOI] [PubMed] [Google Scholar]