Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway (original) (raw)
. Author manuscript; available in PMC: 2012 Oct 28.
Summary
Although infections with virulent pathogens often induce a strong inflammatory reaction, what drives the increased immune response to pathogens compared to non-pathogenic microbes is poorly understood. One possibility is that the immune system senses the level of threat from a microorganism and augments the response accordingly. Here, focussing on cytotoxic necrotizing factor 1 (CNF1), an _Escherichia coli_-derived effector molecule, we showed the host indirectly sensed the pathogen by monitoring for the effector that modified RhoGTPases. CNF1 modified Rac2, which then interacted with the innate immune adaptors IMD and Rip1-Rip2 in flies and mammalian cells, respectively to drive an immune response. This response was protective and increased the ability of the host to restrict pathogen growth, thus defining a mechanism of effector-triggered immunity that contributes to how metazoans defend against microbes with pathogenic potential.
Introduction
Although infections with virulent pathogens are often associated with a strong inflammatory reaction, what drives this increased immune response to pathogens but not commensals is poorly understood. The heightened immune response is often considered to be an epiphenomenon of the increased bacterial load or to be required for the virulence of the invading microbe. An alternative, more host-centric view is that the immune system may be able to sense the level of threat from a microorganism and augment the defense reaction accordingly. However, how virulence is recognized at a molecular level and what strategies contribute to fine-tune the innate immune response after infection with micro-organisms that have pathogenic potential remains poorly understood. What is clear is that models of innate immunity focused on pattern recognition receptors (PRRs) that recognize invariant molecules encoded by microbes (Akira et al., 2001) cannot adequately explain how we discriminate between avirulent and virulent microorganisms as the microbial associated molecular patterns (MAMPs) that are recognized are often intrinsic to all bacteria (Iwasaki and Medzhitov,; Medzhitov,; Vance et al., 2009).
One defining characteristic of pathogenic bacteria is the expression of virulence factors, also called ‘effectors’. These bacterial proteins enter host cells and manipulate host components to promote infection (Galan, 2009) (Ribet and Cossart). One potential strategy for pathogen discrimination is that the host monitors for effectors that are specifically encoded by virulent bacteria and uses this information to gauge the pathogenic potential of the invading organisms. Work in plants has shown this to be the case and, in addition to PRR-based recognition, plants have sophisticated systems of effector-triggered immunity (ETI) based on the direct or indirect recognition of microbial-encoded effectors (Jones and Dangl, 2006). Importantly, this effector-triggered immunity is a powerful means of augmenting the defense response specifically to pathogens but not harmless commensals and makes a significant contribution to how plants restrict pathogen growth. However, although attractive as a strategy of immune surveillance, evidence of how effector-triggered immunity contributes to defense in metazoans is lacking.
A common virulence strategy used by pathogens is to express effectors that target the master regulators of the cytoskeleton, the RhoGTPases (Aktories et al., 2000; Lemonnier et al., 2007). These important proteins are targeted by pathogens because they participate in a number of innate immune processes (Bokoch, 2005). Cytotoxic necrotizing factor 1 (CNF1), produced by uropathogenic Escherichia coli (UPEC), is a prototype of the RhoGTPases-activating toxins (Flatau et al., 1997; Lerm et al., 1999; Schmidt et al., 1997). CNF1 secreted by UPECs is internalized via receptor-mediated endocytosis before translocating into the cytosol of the host cell where the deamidase activity modifies RhoGTPases (Lemmonier et al, 2007). Here we refer to CNF1 as an ‘effector’ as it shares with toxins introduced into the cytosol by protein secretion systems (Galan, 2009) both an intracellular site of action and the ability to induce post-translational modifications of host proteins. Bacteria expressing CNF1 (and the related toxins CNFy from Yersinia pseudotuberculosis and dermonecrotic toxin (DNT) from Bordetella) provoke florid inflammation leading to the suggestion that CNF1 might directly contribute to immune activation. Although previous studies have shown that purified recombinant CNF1 can induce NF-κB activation and cytokine secretion (Munro et al., 2004; Real et al., 2007), the molecular mechanisms that drive the inflammatory response associated with bacteria expressing CNF1 is poorly understood. Similarly, the consequences that this has for both host and pathogen remain to be fully defined.
In Drosophila, two well-defined immune signaling cascades, the Toll and IMD pathways, regulate secretion of a number of antimicrobial peptides (AMPs) such as Drosomycin, acting predominantly downstream of Toll, and Diptericin, being produced as a consequence of IMD activation (Ferrandon et al., 2007; Lemaitre and Hoffmann, 2007). The evolutionarily conservation with mammals has made Drosophila is a useful model for studies of pathogenesis. Interestingly, the insect-pathogenic bacterium Photorhabdus luminescens expresses an effector called Pnf with sequence similarity to the catalytic domain of CNF1 (Waterfield et al., 2002) suggesting that Drosophila might be an appropriate model host in which to study the consequences of this type of effector on host immunity. Here we have taken advantage of tractability of flies to demonstrate the intrinsic immunostimulatory capacity of CNF1 in vitro and in vivo. We found that CNF1 was sufficient to induce an immune response in the absence of other microbial-derived innate immune agonists. This immune response was not simply a bystander of the virulence activity of CNF1 but was protective and conferred upon the host an increased resistance to infection with both CNF1-expressing E. coli and other pathogenic strains. The immune response to CNF1 was initiated not by direct recognition, but indirectly, in response to modification and activation of a host protein, the RhoGTPase Rac2. Modified Rac2 then engaged immune signaling pathways via the innate immune adaptor IMD. Extending these observations to mammals, we showed that this immune response to CNF1 was conserved and triggered by the engagement of Rip proteins after human Rac2 activation. These data defined a mechanism by which the immune system can sense a pathogen not through direct recognition of MAMPs by PRRs but indirectly by sensing the activity of microbial-derived effectors. Importantly, the responses to such effectors can be sufficient to drive protective immunity and help the metazoan host resist microbes with pathogenic potential.
Results
CNF1 induces an effector-triggered immune response that contributes to host resistance
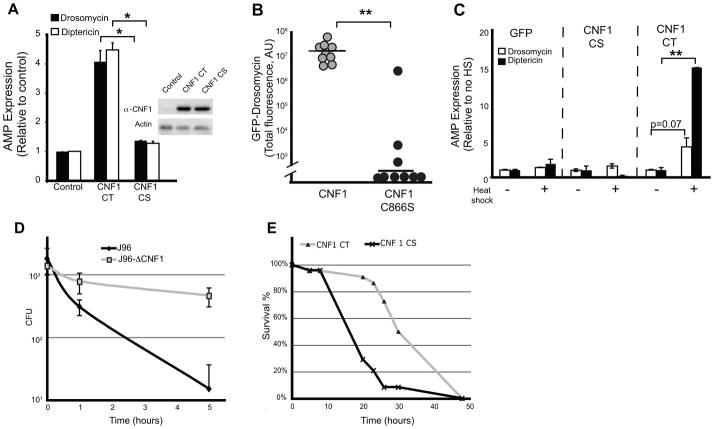
To test if CNF1 directly causes immune activation and if the immunostimulatory capacity was conserved in flies, we expressed CNF1 in Drosophila S2 cells and determined the amounts of induced anti-microbial peptides (AMPs). Consistent with an intrinsic immunostimulatory capacity, expression of CNF1 in S2 cells induced transcriptional upregulation of AMPs (Figure 1A). The immunostimulation required the catalytic activity as expression of the inactive point mutant, CNF1C866S, did not induce AMPs in Drosophila S2 cells. To test if there was immunostimulatory capacity in vivo we injected Drosomycin-GFP reporter flies (Ferrandon et al., 1998) with purified CNF1 or CNF1C866S and assayed the induction of GFP as a surrogate of AMP expression (Figure 1B). Injection of CNF1, but not the catalytically inactive mutant toxin, was associated with induction of AMPs suggesting that the immunostimulatory capacity of CNF1 occurred in vivo. We further generated transgenic expressing either the carboxy-terminus catalytically active CNF1 (CNF1CT) or the catalytically inactive point mutant (CNF1CS) under the control of Upstream Activating Sequences (UAS) allowing us to control expression by the co-expression of GAL4. To test if the effector would be associated with immunostimulation in vivo flies were generated in which CNF1 could be ubiquitously expressed under the control of a temperature sensitive and inducible promoter (HSP70-GAL4; Tubulin-GAL80ts). Similar to our in vitro observations, induction of CNF1CT expression, but not the inactive, CNF1CS, was sufficient to induce AMP expression in vivo (Figure 1C). Together these data indicate that CNF1 has intrinsic immunostimulatory capacity and was sufficient to induce an effector-triggered immune response both in vitro and in vivo.
Figure 1. _E. coli_-derived CNF1 induces effector-triggered immunity in the absence of other bacterial components and contributes to host resistance in vivo.
(A) In vitro induction of Drosomycin (black bars) and Diptericin (white bars) after transfection of catalytically active C-terminus of CNF1 (CNF1CT) and the inactive point mutant (CNF1CS) in Drosophila S2 cells. Insert shows equivalent expression of WT and mutant toxin C-termini. (B) In vivo induction of _Drosomycin_-GFP in reporter flies injected with purified CNF1 (10−5M) or inactive point mutant C866S (CNF1C866S) control toxin. Data expressed as integrated total GFP fluorescence per fly. (C) In vivo induction of AMPs of by heat shock (HS) in flies expressing UAS-GFP or the catalytically active C-terminus of CNF1 (CNF1CT) and inactive point mutant (CNF1CS) under the control of an HSP70-Gal4, tubulin-Gal80ts inducible driver. Data representative of mean+/− s.d. of individual flies in one experiment and representative of experiments using 3 or more independent CNF1CT or CNF1CS transgenic insertion lines. (D) In vivo bacterial loads in OR flies infected with E. coli J96 (black) or mutants lacking CNF1 (grey) at the indicated times. Mean+/− range of c.f.u. per fly calculated at two different dilutions. Data representative of >3 similar experiments. (E) In vivo survival of CNF1CT or control CNF1CS flies infected with Pseudomonas PA14. *p<0.05, **p<0.01.
To determine the in vivo consequences of this effector-triggered immune response we next infected flies with J96 UPEC strain or J96-ΔCNF1 by septic injury and then monitored the persistence of the bacteria in vivo. Expression of CNF1 resulted in rapid eradication of the WT parent strain (J96) whereas deletion of this effector allowed J96-ΔCNF1 bacteria to persist in the host (Figure 1D). Thus expression of CNF1 appeared to favor the host by increasing the ability to eradicate infections with E. coli J96. Pseudomonas aeruginosa PA14 is highly virulent rapidly killing WT flies within hours of septic injury. To test whether the effector-triggered response induced by CNF1 could be protective against a highly virulent microbe we used flies expressing UAS-CNF1CT, or the control UAS-CNF1CS, and infected them with Pseudomonas by septic injury (Figure 1E). Control flies died rapidly reaching 50% mortality by 15 hours. In contrast, UAS-CNF1CT flies showed increased survival, with only 10% death at the same time point and reaching 50% mortality after 30 hours. Thus, CNF1 can function as an ‘avirulence’ factor as its expression drives a protective immune response and increases the ability of the resistant host to clear infections.
CNF1 modifies the Rho-GTPase Rac2, which induces a defense response
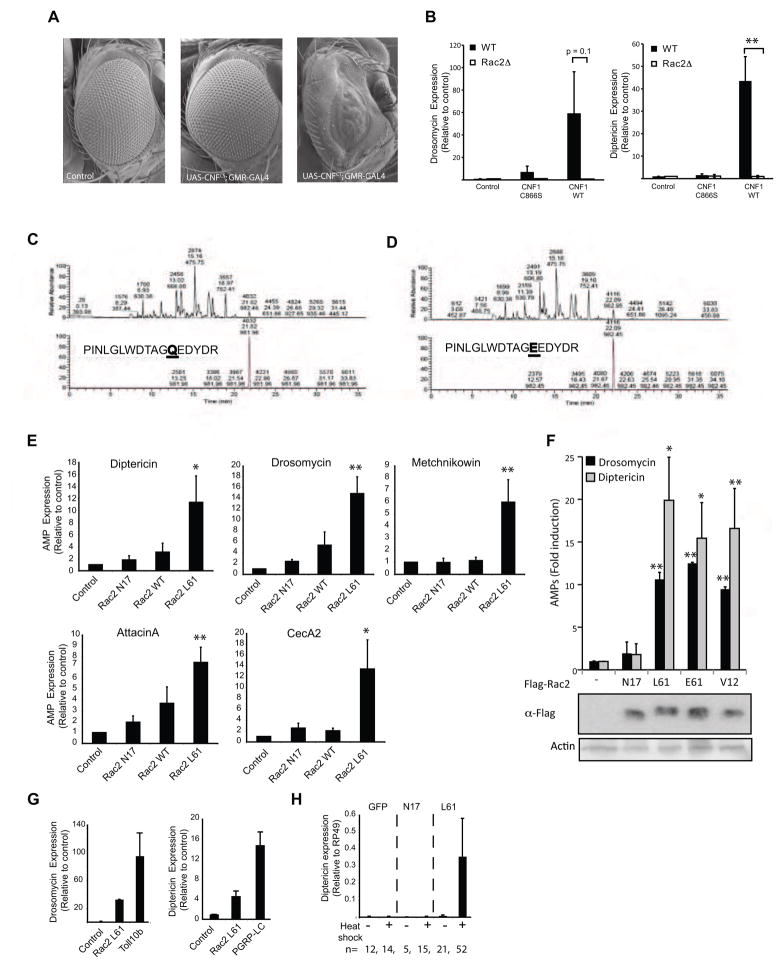
Both heat inactivation and mutation of the catalytic site destroyed the immunostimulatory capacity of CNF1, indicating that the enzymatic activity of CNF1 was required for induction of the Drosophila immune response. In mammals, CNF1 catalyzes a deamidation of the glutamine 63 of Rho or the equivalent 61 of Rac and Cdc42, into a glutamic acid (Flatau et al., 1997; Lerm et al., 1999; Schmidt et al., 1997). This modification abolishes the GTPase activity of the Rho proteins, locking them in a permanent active GTP bound form. We therefore determined if similar CNF1-mediated modification of Rho proteins occurred in flies and was involved in immune activation. Ectopic expression of Rho GTPase in the Drosophila eye has been shown to cause characteristic changes in the ommatidia (Raymond et al., 2001). To confirm that CNF1 was active in flies and to determine which RhoGTPase was likely to be the major in vivo target, we expressed CNF1 in the eye using the GMR-GAL4 driver. Expression of the catalytically active CNF1CT, but not the inactive point mutant CNF1CS, during development was associated with marked disruption of ommatidia and a severe rough eye phenotype confirming the functionality of this toxin in vivo (Figure 2A). Although overexpression-induced eye phenotypes often rely on pleiotropic effects, the phenotype of the GMR-Gal4>UAS-CNF1 flies most closely resembled Rac overexpression, and was distinct from the phenotypes observed after overexpressing of Rho or Cdc42 (Raymond et al., 2001), suggesting that Rac was likely to be the major in vivo target of this effector. To directly test whether the immune response triggered by CNF1 might be mediated by Rac2 we intoxicated Rac2 deficient (Rac2Δ) flies using purified CNF1. AMP expression was abrogated in Rac2Δ flies (Figure 2B) indicating that Rac2 played a non-redundant role in triggering the immune response to this effector. Further suggesting a potential role for Rac2, both J96 and J96-ΔCNF1 showed comparable growth in flies lacking Rac2 (Rac2Δ) (Supplementary Figure SF1a), an observation that was in keeping with the possibility that the immune response triggered by CNF1 was mediated by Rac2. However, consistent with previous reports (Avet-Rochex et al., 2007), and independent of CNF1, Rac2Δ flies showed increased susceptibility to infection with J96 (Supplementary Figure SF1b) and other pathogens and somewhat confounded interpretation of these data.
Figure 2. CNF1 modifies the small RhoGTPase, Rac2, which induces a defense response.
(A) Absence of the ommatidia and a severe rough eye phenotype due to ectopic expression of the toxin in GMR-Gal4/UAS-CNF1CT flies but not control or GMR-Gal4/UAS-CNF1CS flies. (B) In vivo intoxication of WT (black bars) or Rac2Δ (white bars) flies with purified CNF1 toxin and AMPs measured at 6 hours. Drosomycin and Diptericin expression profiles were quantified by QRT-PCR in pools of 3 or more flies (mean+/− s.e.m. of 6 pools from 3 independent experiments). (C & D) Mass spectrometry chromatograms of (C) Native GST-Rac2 and (D) CNF1 modified GST-Rac2. The upper panel represents the total ion chromatogram of GST-Rac2 before (C) or after (D) CNF1 modification. Lower panels show the extracted ion chromatogram for the peptide that includes the position 61 of Rac2 targeted by CNF1 before (C) or after (D) CNF1 modification. The predicted protein sequences are shown. (E) In vitro expression of Diptericin, Drosomycin, Metchnikowin, AttacinA, CecropinA in S2 cells 16h after transfection with Flag-Rac2, Flag-Rac2L61, Flag-Rac2N17 or Flag plasmid (control). AMPs were monitored using QRT-PCR, normalized to RP49 and expressed relative to the empty flag construct (mean+/− s.d.). (F) The upper panel represents the in vitro expression of Diptericin and Drosomycin, in S2 cells 16h after transfection with Flag-Rac2N17, Flag-Rac2L61, Flag-Rac2E61, Flag-Rac2V12. The lower panel is an immunoblot analysis showing levels of expression of the Flag-Rac2 mutants (G) In vitro AMP expression induced by transfection of S2 cells with Rac2L61, Toll10b and PGRP-LC (mean+/− s.d.). (H) In vivo induction of AMPs of by heat shock (HS) in flies expressing UAS-GFP or active Rac2 (Rac2L61) and inactive mutant (Rac2N17) under the control of an HSP70-Gal4, tubulin-Gal80ts inducible driver. Data represent the mean+/− s.e.m. of ‘n’ individual flies pooled from 3 or more independent experiments and representative of results using 3 or more independent Rac2L61 or Rac2N17 transgenic insertion lines. *p<0.05, **p<0.01
To confirm that Drosophila Rac2 could be modified by the catalytic activity of CNF1, GST-purified Rac2 was exposed to CNF1 in a cell-free system and the ability of the toxin to modify Rac2 assessed by mass spectrometry (Figures 2C & D). Purified toxin was sufficient to induce a post-translational modification of the glutamine at amino acid 61 in Drosophila Rac2 (Figure 2D), demonstrating that the activity of this effector was conserved across species and suggesting Rac2 as a likely in vivo target. As mutating amino acid 61 of mammalian Rac results in activation, we next tested if activating mutants of Drosophila Rac2 were associated with AMP induction. Similar to mammalian RhoGTPase, modifying glutamine 61 of Drosophila Rac2 by genetically mutating it to leucine (Rac2L61) was associated with marked cytoskeletal changes and prominent lamellipodia formation (Supplementary Figure SF1c) confirming that modifying this amino acid also resulted in an activating mutant in flies. Expression of Rac2L61, but not the inactive Rac2 mutant (Rac2N17), was sufficient to induce AMP expression in S2 cells and hence recapitulated the effects of CNF1 (Figure 2E). Similar results were seen when two other activating mutants of Rac2 were expressed; the Rac2E61 mutant, mimicking the exact modification induced by CNF1, and Rac2V12 (Figure 2F). Although not as potent as activation of the two innate immune pathways by expression of the receptors Toll10b or PGRP-LC, Rac2L61 induced AMPs to levels 25–30% of these well-studied immune triggers (Figure 2G). Similar to our in vivo observations using CNF1CT transgenic flies (Figure 1C), AMP expression was also upregulated in flies when Rac2L61, but not the inactive mutant, was inducibly expressed in vivo using a heat shock driver (Figure 2H). Together these data indicate that activation of Rac2 is both necessary and sufficient for CNF1 to induce AMP expression.
CNF1-mediated immunostimulation is not due to cytoskeletal disruption
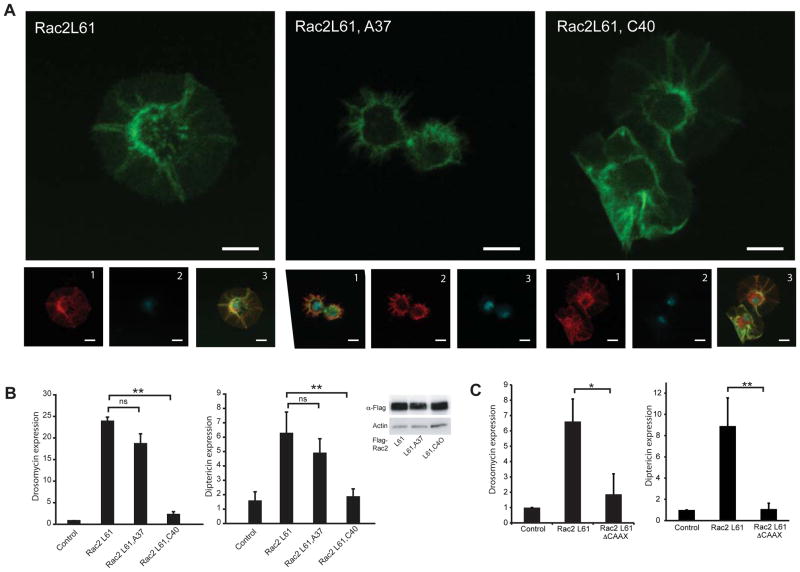
Both Rac2L61 (Supplementary Figure SF1) and CNF1 (Supplementary Figure SF2) expression are associated with marked cytoskeletal changes and prominent lamellipodia formation. Upon activation of RhoGTPase, conformational changes occur in two conserved nucleotide contact regions, switch 1 and switch 2, to allow interaction with the different effector proteins that regulate different downstream signals including cytoskeletal rearrangement. To determine if the effects seen by expression of Rac2L61 was simply due to disruption of the actin cytoskeleton, effector loop mutants of activated Rac2L61 were generated and used to test whether the elicited cytoskeletal changes could be dissociated from the induction of AMPs. Introduction of a F37A mutation into the Rac2L61 blocked lamellipodia formation (Figure 3A) whereas introduction of a Y40C mutation did not affect the regulation of the cytoskeleton (Figure 3A) but disrupted binding to GST-Pak (Supplementary Figure SF2). Similar to Rac2L61, transient transfection of Rac2L61,A37 induced AMP expression. This contrasted with expression of Rac2L61,C40 which was not associated with induction of AMPs (Figure 3B) despite causing prominent cytoskeletal changes. In addition, Rac2L61 had to be able to associate with membranes as the Rac2L61, Delta-CAAX in which the membrane insertion tail had been deleted, was not able to induce AMPs (Figure 3C). Thus signaling to the immune system requires membrane targeting of Rac2 and is regulated through the switch 1 effector-binding domain, providing mechanistic insights into how Rac2 intersects immune signalling pathways. Additionally, these data indicate that Rac2L61 induced a defense response in the absence of other microbial components and independently of its ability to modify of the cytoskeleton.
Figure 3. Rac2 induces an immune response independently of cytoskeletal changes.
(A) Cell morphology of S2 cells expressing Flag-Rac2L61 and switch mutant, Flag-Rac2L61,C40, but not Flag-Rac2L61,A37. Large-scale image is actin (phalloidin-FITC). Smaller images show red, anti-Flag; blue, DAPI and the merge with green, actin (phalloidin-FITC). Scale bar 10μm. (B) In vitro AMP expression in S2 cells expressing Flag-Rac2L61 and switch mutant, Flag-Rac2L61,A37, but not Flag-Rac2L61,C40(mean+/− s.d). Insert demonstrating equivalent expression of all constructs. (C) In vitro AMP expression induced in S2 cells by expressing Flag-Rac2L61 and the ΔCAAX mutant (mean+/− s.d). *p<0.05, **p<0.01.
Rac2 induces activation of defense pathways through an IMD-dependent mechanism
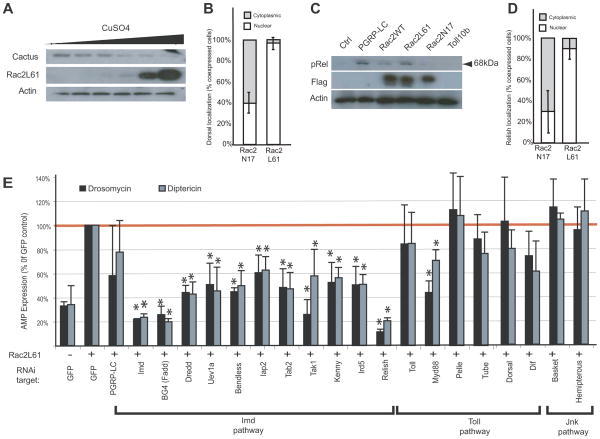
Because Rac2L61 mimicked the molecular changes induced by CNF1, this mutant was used to probe the signaling pathways leading to AMP expression after exposure to this toxin. As disruption of the cytoskeleton did not appear to be essential for Rac2L61 to induce AMP expression, we instead determined if this occurred through any of the known defense pathways in flies. Rac2L61 induced Drosomycin expression (Figure 2), leading us to test whether Rac2L61 activated Dorsal, the NF-kB-like molecule that can regulate this AMP. For Dorsal activation, Cactus, an I-kB-like molecule that acts as an inhibitor of Dorsal by retaining it in the cytoplasm, must be phosphorylated and targeted for proteasomal degradation (Wu and Anderson, 1998). Consistent with Rac2L61-induced degradation of Cactus, increasing the expression of Rac2L61 inversely correlated with the amounts of endogenous Cactus detected (Figure 4A). Correlating with this decrease in Cactus, nuclear localization of GFP-Dorsal occurred in >95% of cells co-transfected with the active Rac2L61 but only 40% with the inactive Rac2N17 mutant (Figure 4B). As Rac2L61 also induced Diptericin, we next determined if it also activated Relish, the NF-κB transcription factor regulating this gene. Activation of Relish (as determined by phosphorylation and cleavage (Erturk-Hasdemir et al., 2009) was detectable when cells expressed Rac2L61 (and the positive control, PGRP-LC) but not Rac2N17 (or the negative control Toll10b) (Figure 4C). Additionally, nuclear translocation of YFP-Relish occurred in >90% of cells co-transfected with the active Rac2 but only 30% co-transfected with the inactive Rac2 mutant (Figure 4D). Thus Rac2L61 was sufficient to activate two of the important innate immune transcription factors, Dorsal and Relish.
Figure 4. Modified Rac2 induces activation of NF-kB-like innate immune transcription factors through an IMD-dependent mechanism.
(A) Immunoblot of Cactus in stable S2 cell lines expressing Rac2L61. Streptavidin–HRP staining indicates levels of BioeaseRac2L61 induced by increasing concentration of CuSO4 as indicated. Actin staining is shown as a loading control. (B) In vitro localization of Dorsal was assayed by blind counting of cells co-transfected with GFP-Dorsal and the Flag-tagged Rac2 mutants, Rac2L61 (active) or Rac2N17 (inactive). (C) Immunoblot showing levels of the transcription factor Relish in S2 cells 16h after transient transfection with Flag-Rac2, Flag-Rac2L61, Flag-Rac2N17, Flag empty plasmid (Ctrl), PGRP-LC or Toll10b plasmids. Flag-tag immunostaining indicates expression of Rac2 proteins. Relish activation was determined by detection of the 68kD phosphorylated, cleaved fragment. Actin staining is shown as a loading control. (D) In vitro localization of Relish localization was assayed by blind counting of cells co-transfected with YFP-Relish and the mRFP tagged Rac2 mutants, Rac2L61 and Rac2N17. (E) Drosomycin (black bars) and Diptericin (grey bars) expression profile were monitored in BioeaseRac2L61 stable S2 cells treated with the indicated RNAi. BioeaseRac2L61 stable cells treated with a GFP RNAi without CuSO4 induction and induced with 500μM CuSO4 are used as controls in each experiment. AMPs expression was determined QRT-PCR and normalized to the house-keeping gene, RP49. Results expressed as % of the induced cells treated with GFP RNAi, (mean+/− s.d.) of 3 independent screens. *, p<0.05, student T test.
To further define the innate immune signaling pathway triggered by Rac2L61 an RNAi epistasis screen was performed in which the components of Toll, IMD and JNK pathways were silenced and their requirement for AMP induction by Rac2L61 determined (Supplementary Figure SF3). Knockdown of all genes in the IMD pathway inhibited Rac2L61-induced AMP expression (Figure 4E). Conversely, silencing of the majority of the Toll pathway components did not decrease the AMP induction by Rac2L61 (Figure 4E). The third innate immune pathway in flies, the JNK pathway, was not required for Rac2L61-induced AMP expression as RNAi silencing neither Basket nor Hemipterous effected Rac2L61-induced AMP expression (Figure 4E). Together these data suggest that Rac2 activates an innate immune response that had potential to regulate both Diptericin and Drosomycin and utilizes signaling components primarily from the IMD pathway.
Modified Rac2 interacts with IMD to induce an immune response
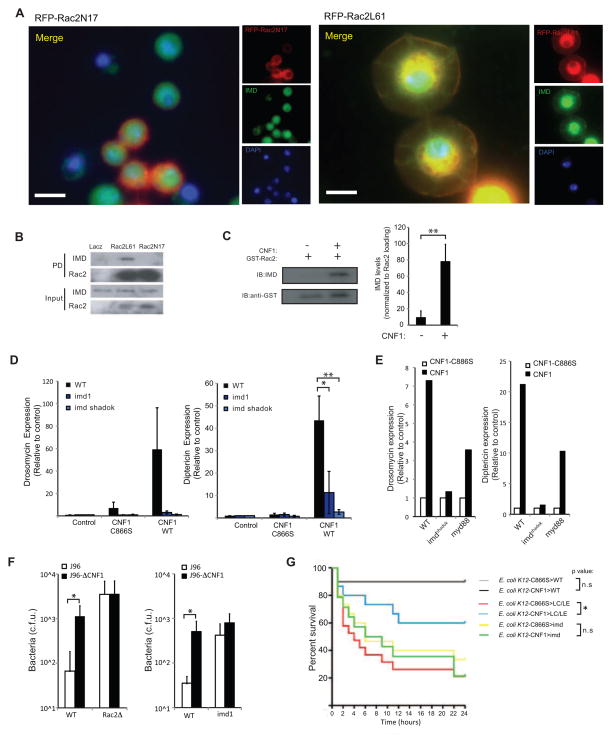
As IMD was the most apical component of the pathway induced by Rac2L61 expression we next tested whether IMD interacted with Rac2. To determine whether these proteins co-localized, mRFP-Rac2L61 or mRFP-Rac2N17 were transiently expressed in S2 cells and their relationship to endogenous IMD determined by microscopy. IMD was detected predominantly in the nucleus both in resting cells (data not shown) and in the presence of mRFP-Rac2N17 (which was found primarily in the cytosol) (Figure 5A). In contrast, when mRFP-Rac2L61 was expressed, a proportion of IMD redistributed and was found in membrane ruffles and in the peri-nuclear region along with Rac2L61 (Figure 5A). A similar redistribution of Flag-IMD was also observed when ectopically expressed with Rac2L61 but not Rac2N17 (Supplementary Figure SF4). To confirm the association of IMD and Rac2 we used S2 cells stably expressing Rac2L61, Rac2N17 or LacZ fused to a Bioease™ tag that allowed intracellular biotinylation of these proteins. IMD was detected in total cell lysates from these three cell lines (Figure 5B). After pulldown of the biotinylated proteins, IMD was found to interact with Rac2L61 but not Rac2N17 or LacZ (Figure 5B). Thus IMD associated biochemically and co-localized by fluorescent microscopy with active Rac2. Using a similar approach we were unable to show any interaction of Rac2L61 with Myd88 (data not shown). The ability of activated Rac2 to bind IMD suggested that CNF1-modified Rac2 might also bind IMD. To test the possibility that this interaction was involved in the induction of AMPs by CNF1 we determined whether Rac2 that had been exposed to CNF1 in a cell-free system had increased affinity for IMD. In GST pull-down assays, native WT Rac2 showed minimal association with IMD whereas Rac2 modified by the toxin CNF1 efficiently bound IMD from cell lysates (Figure 5C). Thus both genetically- and toxin-modified Rac2 interact with the innate immune adaptor, IMD.
Figure 5. Modified Rac2 interacts with the innate immune adaptor IMD to induce AMPs.
(A) Immunofluorescence of S2 cells transfected with mRFP-Rac2L61 or Rac2N17 (red) and colocalization with endogenous IMD (green) determined by antibody staining 16h after transfection. Blue, nuclei (DAPI). (B) IMD interaction with Rac2 were determined by Biotin-Streptavidin based pull-down of biotinylated Rac2 from BioeaseRac2L61 and BioeaseRac2N17 stable S2 cells. BioeaseLacZ acted as a negative control. 2% of total cell lysate before streptavidin purification was loaded on the gel (Input) or proteins eluted after streptavidin pull-down purification (PD). Endogenous IMD was detected by immunostaining. Transfection and pull-down efficiency for the two BioeaseRac2 mutants is shown by Streptavidin–HRP staining of the input and PD specimens respectively. (C) Binding of native GST-Rac2 or GST-Rac2 modified by CNF1 to IMD in S2 cell lysates was determined by immunoblotting. Densitometry of IMD normalized for GST-Rac2 pull-down efficiency in three independent experiments (mean+/− s.d.). (D) In vivo AMPs response to CNF1 was measured in pools of WT (black bars), imd1 (dark blue bars) and _imd_shadok (green bars) flies intoxicated with purified toxin and measured at 6 hours. Drosomycin and Diptericin expression profiles were quantified by QRT-PCR in pools containing 3 or more flies (mean+/− s.e.m. of 6 pools from 3 independent experiments). *p<0.05, **p<0.01. (E) In vivo AMPs response to CNF1 was measured in pools of WT, _imd_shadok and Myd88 flies intoxicated with purified toxin and measured at 6 hours. Drosomycin and Diptericin expression profiles were quantified by QRT-PCR in pools containing 3 or more flies (data representative of 2 similar experiments). (F) In vivo bacterial loads in WT, Rac2Δ (left) or imd1 (right) flies infected with E. coli J96 (white) or mutants lacking CNF1 (black). Mean+/− range of c.f.u. per fly. Data representative of 2 or more similar experiments. (G) Survival or WT, imd1 or PGRP-LC+LE double mutant flies (15–25 per group) infected with E. coli K12-CNF1 or E. coli K12 CNF1-C866S. *p<0.05 using a Gehan-Breslow-Wilcoxon Chi squared Test
We next determined if IMD was required for CNF1 to induce AMPs in vivo using two different IMD mutants, _imd_1, a hypomorphic allele, and _imd_shadok, a null allele. Flies were intoxicated with CNF1 and AMPs determined. Similar to the Rac2Δ mutant flies (Figure 2B), _imd_1 and _imd_shadok, both showed reduced AMP induction after CNF1 exposure (Figure 5D). Consistent with our in vitro observations using RNAi, Myd88 flies showed a partial defect in induction of AMPs after CNF1 intoxication. Notably, the Myd88 flies were not as impaired as the _imd_shadok flies supporting the notion that IMD was the principle adaptor required for this pathway. To confirm that the avirulence activity of CNF1 was indeed propagated by Rac2 and IMD, we infected flies with J96 or J96-ΔCNF1 E. coli and monitored bacterial persistence. Unlike WT flies, which efficiently cleared the J96 strain but not the J96-ΔCNF1 E. coli (Figure 1 and Figure 5F), flies that lacked either Rac2 or IMD were unable to clear either strain, confirming that the avirulence activity required both Rac2 and IMD (Figure 5F and Supplementary figure SF1). Together these data indicate that CNF1 modification of Rac2 and its subsequent interaction with IMD drives the effector-triggered immune response that occurs with this toxin and is required for its avirulence activity.
Finally, we set out to test if CNF1 was sufficient to trigger a protective immune response independently of the two PRRs upstream of IMD, PGRP-LC and PGRP-LE. Because the virulence of J96 was problematic when doing survival experiments with these compromised strains, we instead engineered E. coli in which we could controllably express either CNF1, or the inactive control toxin CNF1-C866S, using IPTG and used these bacteria to infect WT, _imd_1 or PGRP-LC and -LE double mutant flies (Figure 5G). _imd_1 flies succumbed to both bacterial strains whereas WT flies were resistant. In contrast, PGRP-LC and -LE mutant flies were as susceptible as _imd_1 only to E. coli CNF1-C866S but were relatively resistant to _E. coli_-CNF1. These data are consistent with CNF-1 being sufficient to rescue the susceptibility of the PGRP-LC – and -LE mutants by promoting resistance through a PRR-independent, effector-triggered mechanism that relies on IMD
Human Rac2 induces immune activation through Rip1 and Rip2 (RICK)
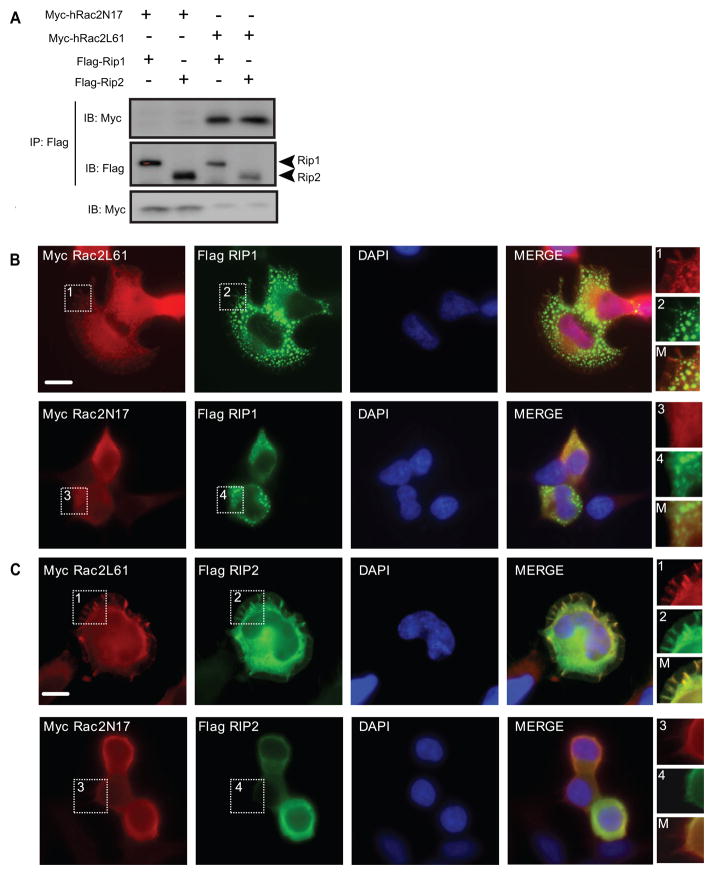
The so-called ‘death’-domains of Drosophila IMD and the mammalian protein Rip1 are homologous. Rip1, and the related molecule Rip2/RICK, are important adaptor proteins for a number of innate immune signaling pathways that function upstream of NF-kB, IRF and caspase-dependent effectors in mammals (Meylan and Tschopp, 2005). We therefore investigated whether, like Drosophila Rac2 and IMD, hRac2 interacted with Rip1 and Rip2. To determine if these proteins interacted Myc-tagged hRac2L61 and hRac2N17 mutants were co-expressed with Flag-tagged Rip1 or Rip2 in HEK-293T cells. hRac2L61, but not hRac2N17, immunoprecipitated with both Rip1 and Rip2 (Figure 6A). To establish the cellular localization of these interactions the proteins were visualized by immunofluorescent microscopy. Although the distribution of staining differed, co-localization of active hRac2L61 was observed with both Rip1 and Rip2 (Figure 6B and 6C). Rip1, which was found in mitochondria (Supplementary Figure SF5), minimally co-localized with hRac2N17 (Figure 6B) but co-localized extensively with Myc-hRac2L61 in these organelles (Figure 6B). Rip2 co-localized with Myc-hRac2L61 in membrane ruffles (Figure 6C).
Figure 6. Modified human Rac2 interacts with the IMD-related molecules, Rip1 and Rip2.
(A) Co-immunoprecipitation of myc-hRac2L61 or hRac2N17 with Flag-Rip1 or Rip2 expressed in HEK293T cells. B & C) Immunofluorescence showing cells co-transfected with a plasmid expressing Myc-tagged hRac2 mutants hRac2L61 or hRac2N17 and the Flag-Rip1 (B) or Flag-Rip2 (C). Red, anti-Myc and green, anti-Flag antibodies. Blue, Nuclei (DAPI). Scale bar, 10μm. High magnification images of indicated fields are shown in inserts.
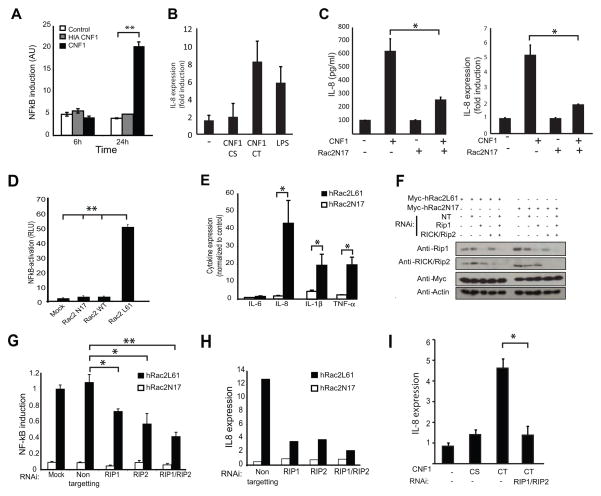
Mammalian epithelial cells do not express many classic PRRs and are relatively resistant to microbial stimuli, a characteristic that is thought to help them tolerate the constant exposure to the harmless commensal microflora. However, the epithelium also remains responsive to pathogen attack, and must therefore utilize alternative non-TLR mechanisms to sense virulent pathogens. As CNF1 is expressed by E. coli J96, a causative agent of urinary tract infection and pyelonephritis in humans, we next set out to determine if human kidney epithelial cells (HEK-293T) might also utilize this type of PRR-independent sensing to respond to uropathogenic bacteria. Consistent with a conserved immunostimulatory capacity and previous reports (Munro et al., 2004), treatment of HEK-293T cells with purified CNF1, but not to heat-inactivated control toxin, was sufficient to induced NF-kB activation in the absence of other microbial components (Figure 7A). The level of induction of the immune response was comparable to that induced by 100ng/ml of LPS (Figure 7B). The ability of CNF1 to induce immune responses required Rac activation as expression of hRac2N17, which can function as a dominant negative inhibitor of Rac, blocked the response (Figure 7C). Furthermore, similar to our observations in Drosophila, expression of active hRac2L61 in HEK-293T cells, but not inactive hRac2N17, caused NF-kB activation (Figure 7D) and transcriptional upregulation of a number of innate immune genes including IL-8, IL-1β, TNFα and type1 interferons as measured by QRT-PCR and RT2 Profiler™ PCR Array (Figure 7E and Supplementary Figure SF6). These data indicate that hRac2L61 had the potential to regulate NF-kB and IRF pathways in mammalian cells and that, similar to flies, Rac2 is both necessary and sufficient for the CNF1 induced immune response.
Figure 7. CNF1 induces an immune response after modification of human Rac2 that induces signaling via IMD-related molecules, Rip1 and Rip2.
(A) NF-kB activation in control HEK 293T cells (white bars), cells exposed to purified heat inactivated CNF1 (grey bars) and CNF1 (black bars) (mean+/− s.d.). (B) IL-8 induction by CNF1-C866S (CS) or CNF1(CT) intoxication or by the addition of 100ng/ml LPS. (C) CNF1 induction of IL-8, after 24h of intoxication, in the presence or absence hRac2N17 that functions as a dominant negative inhibitor of Rac. (D) NF-kB induction and (E) cytokine expression triggered by hRac2 mutants, hRac2L61 or hRac2N17, expression in HEK 293T cells (mean+/− s.e.m., n=3). F-H Rip1 or Rip2 or a Non Targeting (NT) siRNA were transfected in HEK 293T cells 72h as indicated. (F) Endogenous Rip1 and Rip2 immunoblot demonstrates siRNA efficiency and specificity of the siRNA. Anti-Myc staining demonstrates levels of transfection of Myc-hRac2 mutants. Actin staining was used as a loading control. (G) Quantification of hRac2L61 induced NF-kB activation in Rip1 and Rip2 knocked down cells. NF-kB Firefly-luciferase in total cellular lysates was measured and transfection efficiency of the reporter normalized using Renilla activity. Results expressed as % of hRac2L61 expressing cells, mock transfected with siRNA. (H) IL-8 gene expression monitored by QRT-PCR after siRNAi transfection of HEK 293T cells expressing Myc-hRac2 mutants as described above. Actin was used as housekeeping gene and internal control and data expressed normalized to untreated cells. Data is from cells grown in triplicate wells and representative of three independent experiments. (I) CNF1 induction of IL-8, after 24h of intoxication, in the presence or absence RIP1 and RIP2 knock-down. All data are shown as the mean+/− s.d. of biological triplicates and representative of three independent experiments unless otherwise stated. *p<0.05, **p<0.01.
To determine if hRac2 signaled via Rip1 and Rip2 to induce immune activation, HEK-293T cells treated for 48 hours with siRNAs to Rip1, Rip2 or both Rip1 and Rip2 were transfected with hRac2L61 and an NF-kB reporter construct and induction of NF-kB measured after 24h hours. The transfection levels and efficiency of Rip1 and Rip2 silencing by RNAi treatment was confirmed by functional validation and immunoblot (Figure 7F and Supplementary Figure SF6). When compared to non-targeting control siRNA, depletion of Rip1 and Rip2 resulted in a 30% and 40% inhibition of Rac2-dependent NF-kB induction respectively (Figure 7G). Furthermore, the combination of both Rip1 and Rip2 siRNA resulted in 60% inhibition hRac2L61-induced NF-kB activation (Figure 7G). Similar results were observed when IL8 expression was used as a measure of activation and quantified by QRT-PCR (Figure 7H). Similar results were seen with a second, independent siRNA pool (Supplementary Figure SF6). Thus the two mammalian proteins related to IMD, Rip1 and Rip2, also interact with modified Rac2 and contribute to hRac2L61-induced NF-kB activation and IL-8 cytokine expression. Finally, to determine if this was the mechanism by which CNF1 induced immune activation in mammals, we silenced Rip1 and Rip2 in HEK-293T cells and then treated them with CNF1. Paralleling our observations in flies, CNF1 failed to induce an immune response in the absence of Rip1 and Rip2 (Figure 7I), indicating that these proteins played an equivalent role as IMD in the mammalian system. Together these data identify an evolutionary conserved mechanism of effector-triggered immunity triggered by CNF1 modification of the RhoGTPase Rac2, and involving the adaptor proteins IMD or Rip1 and Rip2, in flies and mammalian cell line, respectively.
Discussion
Here, we have focused on Escherichia coli toxin CNF1 as a prototype of bacterial effectors that activate the RhoGTPases (Aktories et al., 2000; Lemonnier et al., 2007), and have demonstrated that this toxin is sufficient to induce an immune response in the absence of other microbial-derived innate immune agonists. Using Drosophila as a genetically tractable system, we have found that this effector-triggered immune response is initiated not by direct recognition of CNF1, but in response to modification and activation of a host protein, Rac2, which then engages immune signaling pathways primarily via the innate immune adaptor IMD. Extending these observations to mammalian cells, we show that the immune response to CNF1 is conserved and triggered by the engagement of Rip proteins by activated human Rac2. The precise molecular mechanism of how the immune response is activated by Rac2 will need to be further defined. However, our observation that deletion of the CAAX-box, which is necessary for membrane targeting of RhoGTPases, prevents Rac2L61-induced AMPs leads us to propose that Rac2 functions by stabilizing the interaction of signaling adaptors at such sites and can therefore obviate the need for engagement of PRRs. This possibility is further supported by data in mammals in which it has been shown that membrane targeting of RIP2 by addition of a myristolation sequence is sufficient to induce NF-κB activation and bypasses the requirement for the receptor NOD2 (Lecine et al., 2007).
Reminiscent of effector-triggered immunity in plants, which has been shown to be critical for host defense in resistant strains (Dangl and Jones, 2001; Jones and Dangl, 2006), the immune response to CNF1 is not simply a bystander of the virulence activity but can confer upon the host resistance to infection. Thus, this mechanism of pathogen surveillance provides an example of how immunity in metazoans can be triggered independently of MAMP recognition by PRRs. Importantly, this effector-triggered immune response occurs when microbes express so-called ‘virulence’ factors and hence is likely to be induced specifically during infection with bacteria with pathogenic potential.
A number of models have been suggested to explain how the immune system specifically amplifies the response to pathogens but not harmless commensals. As an example, one proposed mechanism of sensing of virulent microbes is through the NLRP3 inflammasome, which can be activated by pathogens that express pore forming toxins and cause membrane disruption (Freche et al., 2007; Gurcel et al., 2006; Mariathasan et al., 2006). However, other studies attempting to identify how the host senses and responds to the secretory apparatus and/or effectors of bacteria have concluded that the responses are mediated by non-canonical immune pathways (Auerbuch et al., 2009; Bruno et al., 2009; Shin et al., 2008). Similarly, the response to CNF1 described herein does not require NLRs, which are absent from the innate immune arsenal of Drosophila, indicating that indirect sensing of virulent microbes does not rely solely on the inflammasome and can be mediated by other immune pathways. Although we would draw comparisons between our system and any other pathogen with caution, together these studies point to the existence of alternative, but currently poorly defined, mechanism of immune activation that allow the host to differentiate virulent from avirulent microorganisms. Our data are more consistent with a second model of pathogen recognition that suggests that the host responds to the associated cellular and tissue damage (Matzinger, 1994; Matzinger, 2002; Casadevall and Pirofski, 2003). These ideas have recently been included in a more expansive framework of pathogen vs. non-pathogen discrimination that takes into consideration these and other features that are associated specifically with virulent microbes termed ‘Patterns of Pathogenesis’ (Vance et al., 2009). Our findings expand this framework to include very specific perturbations of homeostasis caused when effectors target critical signalling pathways or essential regulatory proteins.
Recent work has highlighted the complex strategies evolved by microbes during the course of the host-pathogen dynamic and indicate that not all bacterial effectors can be considered as simple virulence factors that act solely to the detriment of the host. Indeed, many pathogens have evolved bacterial effectors that modify the host cell cytotoxicity (Shames and Finlay). Not only can effectors limit damage but previous work has shown that they can also drive immune responses both in vitro (Bruno et al., 2009) and in vivo (Gottar et al., 2006). However, the in vivo consequences of the increased defense reaction remains to be defined and, in most cases, it is assumed to be part of the virulence strategy of the microbe or due to the increased bacterial load. This contrast with the situation for CNF1, which we show is sufficient to induce protective immunity. This important distinction indicates that the response to CNF1 has similar consequences to the effector-triggered immunity described in plants, where it is well recognized to act to limit microbial replication and protect a resistant host. Historically, investigators in the plant field have referred to these sorts of effectors as ‘avirulence’ factors as, when expressed by bacteria, they negatively impact the ability of a pathogen to infect the host (Mansfield, 2009). However, it is now understood in the plant system that such effectors are directly or indirectly recognized by the host to initiate effector-triggered immunity and hence by augmenting the immune response contribute to host resistance to the microbe. These observations do raise the question as to why pathogens would retain such ‘avirulence’ genes, a point that has been debated by plant immunologists for years. However, it is now clear from the work in plants that not all hosts mount a response and responsiveness depends on whether a host has the correct receptor. In a resistant host the effector acts as an ‘avirulence gene’ whereas in hosts that lack the receptor, the effector promotes virulence. Thus it is a tradeoff for the pathogen as the factor that allows it to infect one host may also be the thing that prevents it effectively infecting another. This variation in ability to respond to an effector may only be apparent at a population/species level. Alternatively, in the context of a complex metazoan such as flies or mammals, it may be different in different tissues. Thus variation in resistance and susceptibility due to a differential ability to mount an ETI may explain the tropism of certain pathogens for certain tissues and/or the variation in host susceptibility across the population, both possibilities that will need to be further explored. The observation that species phylogenetically distant from plants also monitor and respond to microbial effectors suggest that this is a convergent evolutionary strategy for identifying and defending against virulent pathogens. Taken together these data define a mechanism of effector-triggered immunity and show how it can be a potentially important means by which metazoans discriminate harmless microbes from those with pathogenic potential and differentially respond to virulent microorganisms.
Methods
Standard methods and methods pertaining to the mammalian work can be found in the supplement.
Fly infections, intoxication and survival experiments
For details of fly stocks please see supplementary methods. UAS-CNF1 lines were maintained at 18C and crossed to Pw+mC Tub-Gal80ts2;hsp-Gal4 drivers (gift of D. Ferrandon) to induce toxin expression by heat shock (30 min 37°C, 30 min 25°C, 30 min 37°C, and 16 hr incubation 25°C). For CNF1CT and CNF1CS flies AMPs profile analysis, single fly qRT-PCR was performed. Expression of the transgene was confirmed by PCR. Three or more independent lines with different insertions were analyzed.
E. coli J96 or J96-ΔCNF1 were grown in LB overnight at 37°C and diluted ¼ and subcultured up to OD600= 0.7. Cultures were centrifuged (3000rpm, 5min) and the supernatant was removed. A tungsten needle was dipped into the bacterial pellet and used to prick 15–25 adult male flies that had been placed on sterile food for 24 hours previously. Pools of 3 flies were homogenized in PBS and dilutions plated and grown on Streptomycin plates overnight. Bacterial loads were determined by colony counting. For some survival experiments E. coli K12 were transformed with pQE30-His-CNF1 or pQE30-His-CNF1-C866S. Bacteria were grown overnight at 37°C in LB supplemented with ampicilin (100μg/ml) and IPTG (10mM). Bacteria were then diluted 1/100 and subcultured up to OD600= 0.8 and then IPTG (1mM) was added for 16h at 30°C. A tungsten needle was dipped into the bacterial pellet and used to prick 15–25 adult flies. Flies were then left in a sterile sucrose 5% supplemented with (1mM) IPTG and mortality was monitored for 24h. Flies were intoxicated either by micro-injection with 65nl of 10−5M recombinant purified CNF1 toxin using Nanoject microinjector or by placing in vials containing a sucrose-based solution (5% sucrose, 10mM Tris [pH7.4] containing 3.5mg/ml of purified recombinant CNF1 toxin or the catalytically inactive CNF1 mutant, C866S. Control flies were added to vials containing only sucrose. After 6 hours flies were lyzed and processed for Q-RT-PCR. Three or more pools of three to five age matched flies (males only) from WT, Rac2Δ, imd1 or imdshadok were used.
RNA interference and epistasis analysis in Drosophila S2 cells
For screening, S2 cells stably expressing BioeaseRac2L61 (S2BioeaseRac2L61) were used as they demonstrated a reproducible 3-fold induction of AMPs after CuSO4 induction (Supplementary Figure SF3). Double-stranded RNA were synthesized and purified as described previously (Stuart et al., 2007). Primers were designed using sequences from the Drosophila Research Screening Center (Supplementary Table 1). The efficacy of the RNAi collection used was confirmed using cells expressing either PGRP-LC, Toll10b or stimulated with PGN (Supplementary Figure SF3). All RNAis were functional, although Dif and Dorsal are known to be partially redundant in this S2 system (Silverman et al., 2000). Cells were treated with RNAi 72 hours prior induction of BioeaseRac2L61 expression for 16 hours, expression of both Drosomycin and Diptericin was compared to CuSO4 induced cells treated with a control non-targeting RNAi.
Supplementary Material
01
Highlights.
- The bacterial effector CNF1 is sufficient to drive a protective immune response
- CNF1 modification of Rac2 triggers antimicrobial peptide response
- Activation of Rac2 triggers an immune response via the IMD signaling pathway
- Human Rac2 induces immune activation through Rip1 and Rip2
Acknowledgments
We would like to thank D. Ferrandon, J.L. Imler and C. Kocks for sharing fly stocks, K. Fitzgerald, J. Tschopp, G. Gacon and the Drosophila Genomics Resource Center for plasmids and E. Lemichez and G. Flatau for providing CNF1 and CNF1 mutant purified proteins and cDNA and anti-CNF1 antibody. We thank J. Dejardin, O. Visvikis, P. Leopold, M. Maddugoda, A. Mettouchi, B. Ferrua, P. Munro and A. Doye for technical suggestions and F. Ausubel, A Ezekowitz and E. Lemichez for critical discussion and reading the manuscript. Technical support was provided by the C3M Cell Imaging Facility (Francois Prodon) and the Taplin Mass Spectrometric Facility (Ross Tomaino). L.B. was supported by Fondation Recherche Medical, MGH ECOR Fund for Medical Discovery and Ligue Nationale Contre le Cancer. This work was supported by NIH/NIAID grants to J-M.R. (PO1: A104420) and N.S (RO1: A160025) and grants to L.S from MGH ECOR and NIH/NIAID.
Footnotes
Author Contributions
All experiments were designed and interpreted by LB and LMS. LMS supervised the work with assistance from ALH. LB and LMS wrote the manuscript with input from ALH and NS_. In vitro_ experiments were performed by LB, SD, CH, GMC, EH and WKI. Flies were generated and maintained by LB, MC and LM. In vivo experiments were performed by LB, LM, MC, NP and SF. IMD, Relish, Dorsal and PGRP-LC reagents (constructs antibodies etc) were generated and supplied by DEH, JMR and NS
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Aktories K, Schmidt G, Just I. Rho GTPases as targets of bacterial protein toxins. Biol Chem. 2000;381:421–426. doi: 10.1515/BC.2000.054. [DOI] [PubMed] [Google Scholar]
- Auerbuch V, Golenbock DT, Isberg RR. Innate immune recognition of Yersinia pseudotuberculosis type III secretion. PLoS Pathog. 2009;5:e1000686. doi: 10.1371/journal.ppat.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avet-Rochex A, Perrin J, Bergeret E, Fauvarque MO. Rac2 is a major actor of Drosophila resistance to Pseudomonas aeruginosa acting in phagocytic cells. Genes Cells. 2007;12:1193–1204. doi: 10.1111/j.1365-2443.2007.01121.x. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 2005;15:163–171. doi: 10.1016/j.tcb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galan JE. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. The damage-response framework of microbial pathogenesis. Nat Rev Microbiol. 2003;1:17–24. doi: 10.1038/nrmicro732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Erturk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stoven S, Meier P, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci U S A. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann JA. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. Embo J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- Freche B, Reig N, van der Goot FG. The role of the inflammasome in cellular responses to toxins and bacterial effectors. Semin Immunopathol. 2007;29:249–260. doi: 10.1007/s00281-007-0085-0. [DOI] [PubMed] [Google Scholar]
- Galan JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, Belvin M, Hoffmann JA, Ferrandon D. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Lecine P, Esmiol S, Metais JY, Nicoletti C, Nourry C, McDonald C, Nunez G, Hugot JP, Borg JP, Ollendorff V. The NOD2-RICK complex signals from the plasma membrane. J Biol Chem. 2007;282:15197–15207. doi: 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lemonnier M, Landraud L, Lemichez E. Rho GTPase-activating bacterial toxins: from bacterial virulence regulation to eukaryotic cell biology. FEMS Microbiol Rev. 2007;31:515–534. doi: 10.1111/j.1574-6976.2007.00078.x. [DOI] [PubMed] [Google Scholar]
- Lerm M, Selzer J, Hoffmeyer A, Rapp UR, Aktories K, Schmidt G. Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1: activation of c-Jun N-terminal kinase in HeLa cells. Infect Immun. 1999;67:496–503. doi: 10.1128/iai.67.2.496-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JW. From bacterial avirulence genes to effector functions via the hrp delivery system: an overview of 25 years of progress in our understanding of plant innate immunity. Mol Plant Pathol. 2009;10:721–734. doi: 10.1111/j.1364-3703.2009.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Innate immunity: quo vadis? Nat Immunol. 11:551–553. doi: 10.1038/ni0710-551. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Munro P, Flatau G, Doye A, Boyer L, Oregioni O, Mege JL, Landraud L, Lemichez E. Activation and proteasomal degradation of rho GTPases by cytotoxic necrotizing factor-1 elicit a controlled inflammatory response. J Biol Chem. 2004;279:35849–35857. doi: 10.1074/jbc.M401580200. [DOI] [PubMed] [Google Scholar]
- Raymond K, Bergeret E, Dagher MC, Breton R, Griffin-Shea R, Fauvarque MO. The Rac GTPase-activating protein RotundRacGAP interferes with Drac1 and Dcdc42 signalling in Drosophila melanogaster. J Biol Chem. 2001;276:35909–35916. doi: 10.1074/jbc.M105779200. [DOI] [PubMed] [Google Scholar]
- Real JM, Munro P, Buisson-Touati C, Lemichez E, Boquet P, Landraud L. Specificity of immunomodulator secretion in urinary samples in response to infection by alpha-hemolysin and CNF1 bearing uropathogenic Escherichia coli. Cytokine. 2007;37:22–25. doi: 10.1016/j.cyto.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- Shames SR, Finlay BB. Breaking the stereotype: virulence factor-mediated protection of host cells in bacterial pathogenesis. PLoS Pathog. :6. doi: 10.1371/journal.ppat.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Case CL, Archer KA, Nogueira CV, Kobayashi KS, Flavell RA, Roy CR, Zamboni DS. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008;4:e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Boulais J, Charriere GM, Hennessy EJ, Brunet S, Jutras I, Goyette G, Rondeau C, Letarte S, Huang H, et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield NR, Daborn PJ, ffrench-Constant RH. Genomic islands in Photorhabdus. Trends Microbiol. 2002;10:541–545. doi: 10.1016/s0966-842x(02)02463-0. [DOI] [PubMed] [Google Scholar]
- Wu LP, Anderson KV. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392:93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01