Structural Basis for Specific Recognition of Rpt1p, an ATPase Subunit of 26 S Proteasome, by Proteasome-dedicated Chaperone Hsm3p (original) (raw)
Background: Hsm3 is a proteasome-dedicated chaperone that forms a base precursor, Hsm3-Rpt1-Rpt2-Rpn1.
Results: Crystal structures of Hsm3 and Hsm3-Rpt1 were determined.
Conclusion: The Hsm3-Rpt1 interface is formed by a hydrophobic core and complementary charged interactions and is important for the proteasome assembly.
Significance: The study provides the structural basis for the assembly mechanism of the base subcomplex of the 26 S proteasome.
Keywords: Chaperone Chaperonin, Crystal Structure, Proteasome, Protein Degradation, Ubiquitin
Abstract
The 26 S proteasome is a 2.5-MDa molecular machine that degrades ubiquitinated proteins in eukaryotic cells. It consists of a proteolytic core particle and two 19 S regulatory particles (RPs) composed of 6 ATPase (Rpt) and 13 non-ATPase (Rpn) subunits. Multiple proteasome-dedicated chaperones facilitate the assembly of the proteasome, but little is known about the detailed mechanisms. Hsm3, a 19 S RP dedicated chaperone, transiently binds to the C-terminal domain of the Rpt1 subunit and forms a tetrameric complex, Hsm3-Rpt1-Rpt2-Rpn1, during maturation of the ATPase ring of 19 S RP. To elucidate the structural basis of Hsm3 function, we determined the crystal structures of Hsm3 and its complex with the C-terminal domain of the Rpt1 subunit (Rpt1C). Hsm3 has a C-shaped structure that consists of 11 HEAT repeats. The structure of the Hsm3-Rpt1C complex revealed that the interacting surface between Hsm3 and Rpt1 is a hydrophobic core and a complementary charged surface. Mutations in the Hsm3-Rpt1 surface resulted in the assembly defect of the 26 S proteasome. Furthermore, a structural model of the Hsm3-Rpt ring complex and an in vitro binding assay suggest that Hsm3 can bind Rpt2 in addition to Rpt1. Collectively, our results provide the structural basis of the molecular functions of Hsm3 for the RP assembly.
Introduction
The 26 S proteasome, a 2.5-MDa multiprotein complex built from at least 33 different subunits, plays a key role in ubiquitin-dependent proteolysis by degrading proteins conjugated to ubiquitin (1–3). This enzyme consists of a proteolytic 20 S core particle (CP)4 and one or two 19 S regulatory particles (RPs), including the six regulatory particle AAA-ATPase (Rpt) and the 13 regulatory particle non-ATPase (Rpn) subunits. The 20 S CP is a cylindrical particle that is formed by axial stacking of four heteroheptameric rings, α1–7β1–7β1–7α1–7. The seven distinct α subunits (α1–7) and seven distinct β subunits (β1–7) of the eukaryotic proteasomes each occupy a unique position in their respective rings. The 19 S RP is responsible for targeting the ubiquitin-tagged substrate, unfolding the substrate protein, activating the CP by opening the gate, and releasing ubiquitin from the substrate. The 19 S RP consists of at least 19 different subunits and can be divided into two subcomplexes, the base and the lid. The base is composed of six different homologous Rpt subunits, Rpt1–6, and three Rpn subunits, Rpn1, Rpn2, and Rpn13. The Rpt subunits are required for substrate unfolding and α-ring channel opening. The lid is composed of nine non-ATPase subunits (Rpn3, Rpn5–9, Rpn11–12, and Rpn15), most of which are essential for viability, but their precise roles are still elusive except for Rpn11, which displays a deubiquitylating activity to spare ubiquitin. The lid-base association is stabilized by the ubiquitin receptor Rpn10 (4, 5).
Recent studies have suggested that multiple proteasome-dedicated chaperones are involved in efficient and correct assembly of the 26 S proteasome (6–8). For the 20 S CP assembly, five distinct chaperones are identified as follows: Ump1 (9, 10); PAC1-PAC2 complex (11) (called Pba1-Pba2 complex (12) in yeast) and PAC3-PAC4 complex (13) (Pba3-Pba4 complex in yeast) (14, 15). These chaperones help the initiation and progression of the assembly process by transiently associating with proteasome precursors. More recently, four specific chaperones, Nas2/p27, Nas6/gankyrin, Rpn14/PAAF1, and Hsm3/S5b, were identified for the base assembly both in yeast and in mammals (16–19). Each base-dedicated chaperone is responsible for the formation of three distinct subassemblies (Nas2/p27-Rpt5-Rpt3, Nas6/gankyrin-Rpt3-Rpt6-Rpn14/PAAF1, and Hsm3/S5b-Rpt1-Rpt2-Rpn1) and escorts them until the 19 S RP is formed. Because these chaperones are not present in the mature 26 S proteasome, these chaperones should be released at certain steps in the assembly process, but the precise mechanism is still unclear.
We previously reported the crystal structures of Pba3-Pba4 complexed with the α5 subunit of the CP (15) and Rpn14 (20). Although these studies provided a structural basis for mechanisms underlying proteasome assembly, detailed mechanisms regarding how the RP is formed remain mostly unclear. Hsm3 is a HEAT (huntingtin-elongation-A subunit-TOR) repeat-containing protein. Previous yeast genetic studies have shown that deletion of Hsm3 shows the most severe phenotypes among the RP-dedicated chaperones, indicating the formation of the tetrameric complex, Hsm3-Rpt1-Rpt2-Rpn1, is critical for the RP assembly (16–19).
In this study, we determined the crystal structures of full-length Hsm3 and the complex with the C-terminal domain of Rpt1 (Rpt1C). We show that a specific interaction between Rpt1 and Hsm3 is required for the proper assembly of the 19 S RP. By a structural modeling of the Rpt ring-Hsm3 complex and in vitro binding assays, we propose the model that Hsm3 has a critical role for proper arrangement of Rpt1 and Rpt2.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification for Crystallizations
We amplified the Saccharomyces cerevisiae Hsm3 open reading frame (ORF) by PCR from yeast genomic DNA of BY4741 and cloned it into the pET28(+) vector (Novagen). We also constructed the pET21-Rpt1C vector, which expresses the C-terminal region (amino acids 381–467) of Rpt1. Site-directed mutants were generated with the QuikChange site-directed mutagenesis kit (Stratagene) using pET28-Hsm3 and pET21-Rpt1C as the template.
The His6-tagged Hsm3 was expressed from pET28a plasmid (Novagen) in Escherichia coli BL21 (DE3). The protein was purified using nickel affinity (Ni-NTA, Millipore), anion exchange (Q-Sepharose, GE Healthcare), and gel filtration (Superdex75, GE Healthcare) chromatographies. The SeMet derivative Hsm3 was prepared similarly using E. coli B834 (DE3) cells grown in minimal medium containing l-selenomethionine (25 μg/ml). The WT and SeMet Hsm3 were then concentrated to 30 mg/ml by ultrafiltration in 25 mm Tris-HCl (pH 7.5) and 1 mm dithiothreitol.
For the co-crystallization of Hsm3 and the Rpt1C complex, two vectors of Hsm3 cloned into pET28a (kanamycin-resistant) and Rpt1C cloned into pET21 (ampicillin-resistant) were co-transformed into E. coli BL21 (DE3), and two proteins were simultaneously expressed by isopropyl 1-thio-β-d-galactopyranoside induction. Hsm3 was expressed as a His6-tagged protein, and Rpt1C was expressed as an untagged protein. After cell lysis, the Hsm3-Rpt1C complex was purified by nickel-affinity, anion exchange, and gel filtration chromatographies. The Hsm3-Rpt1C complex was then concentrated to 80 mg/ml and used for crystallization.
Crystallization and Data Collection
Crystals of WT Hsm3 and SeMet Hsm3 were obtained at 20 °C by using the hanging-drop vapor diffusion method. Although WT and SeMet Hsm3 crystals were grown from 0.1 m Tris-HCl (pH 7.0), 0.2 m KH2PO4, and 8% (w/v) PEG3350, they were not sufficient for crystal structure determination. We obtained single crystals for WT and SeMet Hsm3 under the same crystallization condition by microseeding, which produced two crystal forms. The WT and the SeMet Hsm3-Rpt1C crystals were prepared by using 0.1 m _N_-(2-acetamido)iminodiacetic acid (pH 8.5), 1 m NaCl, 0.8 m Li2SO4, 0.5% (w/v) PEG3350 at 4 °C. Crystals were equilibrated in cryoprotectant buffer containing reservoir buffer plus 26% (v/v) glycerol and then cryogenized in a cold nitrogen stream at 100 K.
Diffraction data sets for WT and SeMet Hsm3 and Hsm3-Rpt1C were collected at beamline BL44XU (SPring8, Hyogo, Japan). Data processing and reduction were carried out with HKL2000 (21). The crystal forms of WT Hsm3, SeMet Hsm3, and Hsm3-Rpt1C belong to the space group _P_21, with cell dimensions of a = 78.8, b = 91.9, c = 78.9 Å, β = 113.1°, _P_212121, with cell dimensions of a = 85.4, b = 94.7, c = 129.2 Å, _P_6522, with cell dimensions of a = b = 187.3, c = 380.3 Å. Each of them contains two molecules in the asymmetric unit, respectively. Data collection statistics are given in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics
Structure Determination and Refinement
The structure of Hsm3 was determined by the multiwavelength anomalous dispersion method using a selenomethionine derivative. The position of the heavy atoms was obtained using SHELXD (22) and refined using MLPHARE (23). Initial multiwavelength anomalous dispersion phase was extended to 2.05 Å resolution with solvent flattening and histogram mapping using DM (24). An initial model was built using ARP/wARP (25). Manual building was then carried out using the program COOT (26) and alternated with several cycles of refinement using the program REFMAC5 (27). The final SeMet Hsm3 model contained residues 9–465 and 9–465 of the molecules in the asymmetric unit (molecules A and B), respectively. The structure of WT Hsm3 was determined by the molecular replacement technique using MOLREP (28) with the refined model of SeMet Hsm3. Structure refinement of the WT Hsm3 was guided by referring to the structure of the wild type. The final refined model consists of residues 11–23, 31–43, 49–64, and 74–466 and residues 13–27, 36–43, 49–64, and 75–465 of the molecules in the asymmetric unit (molecules A and B), respectively.
The structure of Hsm3-Rpt1C was solved by the molecular replacement with the structure of Hsm3 and the C-terminal domain of Rpt3 (Rpt3C) (PDB code 2DZN) using the program MOLREP. Model building was carried out using the program COOT. The electron density map agreed well with that of the Hsm3 molecule. Although it was difficult to trace the Rpt1C because of insufficient quality of the electron density map, the methionine-marking approach successfully revealed the overall structure of Rpt1C. The structure of the Hsm3-Rpt1C complex was refined with REFMAC5 and CNS (29, 30). The refined model contains residues 9–465, 7–420, and 426–465 of Hsm3 (molecules A and C) and residues 380–452 and 380–452 of Rpt1C (molecules B and D), respectively. There are no residues in the disallowed regions of the Ramachandran plot. Phasing and refinement statistics are summarized in Table 1. Structure figures were generated using PyMol (31) and CCP4MG (32).
Binding Assays by Using E. coli Expression System
His6-tagged Hsm3 or mutants were co-expressed with untagged Rpt1C or its mutants in E. coli. A small aliquot (∼100 μl) of Ni-NTA was washed with wash buffer (20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 20 mm imidazole). Clarified lysate (∼5 ml) was added to the resin and incubated at room temperature for 5 min. After incubation, beads were sedimented by centrifugation and washed three times with 1 ml of wash buffer to remove contaminating protein. Finally, the beads were mixed with equal volume of 2× SDS-PAGE loading dye and boiled at 95 °C to release bound proteins from the beads. Protein samples were analyzed by SDS-PAGE, followed by Coomassie Brilliant Blue staining.
Binding Assays by Using a Wheat Germ Cell-free Protein Synthesis
To analyze the interaction between the full-length proteasome subunits and Hsm3, we used a wheat germ cell-free expression system because the system has an advantage for producing a broad range of difficult-to-express proteins in E. coli (33, 34). The ORFs of the proteasome subunits and Hsm3 were cloned into pEU-E01-MCS vector or pEU-E01-His-TEV-MCS vector (CellFree Sciences, Matsuyama, Japan). His6-tagged proteasome subunits and Hsm3 proteins were co-synthesized by the standard bilayer methods according to the manufacturer's instructions for a small scale protein expression (WEPRO7240H, CellFree Sciences, Matsuyama, Japan). After centrifugation for 10 min at 20,000 × g, 2 μl of nickel-Sepharose HP (GE Healthcare) was added to the supernatants (∼200 μl) and incubated for 2 h at 4 °C. After washing three times with wash buffer (50 mm phosphate buffer (pH 7.5), 300 mm NaCl, 50 mm imidazole), the proteins were eluted by 20 μl of elution buffer (50 mm phosphate buffer (pH 7.5), 300 mm NaCl, 500 mm imidazole). The protein samples were analyzed by 12% low-Bis SDS-PAGE, followed by Coomassie Brilliant Blue staining.
Yeast Strains and Media
Standard techniques were used for strain constructions and transformations (35). SC medium consisted of 0.67% yeast nitrogen base without amino acids, 0.5% casamino acids, 2% glucose, 20 mg/liter tryptophan, 20 mg/liter uracil, 400 mg/liter adenine, and 10 mm phosphate buffer (pH 7.5). All strains listed in Table 2 are congenic to W303. To construct rpt1 mutants, DNA fragment encoding 5′-truncated RPT1 (+285 to +1753) was inserted into the YIp vector pRS303 (36), and the residues corresponding to Arg-403 and/or Arg-409 of Rpt1 were mutated to Ala by the QuikChange site-directed mutagenesis kit (Stratagene). The resulting plasmids were linearized at its unique AlfII site (+445) and transformed into the wild-type diploid cells (W303D). The positive transformants, in which correct integration was confirmed by genomic DNA sequencing, were subjected to tetrad analysis to recover haploid rpt1 mutants (YYS1867, YYS1899, and YYS1905). For hsm3 mutants, we constructed the low copy centromeric vector that expresses hsm3 mutant with a HA5 tag under the HSM3 promoter (600 bp) in the YCplac22-based plasmid pTS901CT (37). After DNA sequencing, the plasmids were transformed into the hsm3 deletion strain YYS1204.
TABLE 2.
Yeast strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| W303-1A | MATa ura3-1, trp1-1, leu2-3,112, his3-11,15, ade2-1, can1-100 | Our stock |
| YYS1897 | MATa rpt1::rpt1R403A-HIS3 | This study |
| YYS1899 | MATa rpt1::rpt1R409A-HIS3 | This study |
| YYS1905 | MATa rpt1::rpt1R403A/R409A-HIS3 | This study |
| YYS1204 | MATa Δ_hsm3_::KanMX | 19 |
| YYS1771 | MATa Δ_hsm3_::KanMX [_YCplac22_] | This study |
| YYS1772 | MATa Δ_hsm3_::KanMX [_YCplac22-HSM3pr-HSM3-HA_5] | This study |
| YYS1917 | MATa Δ_hsm3_::KanMX [_YCplac22-HSM3pr-hsm3E157A-HA_5] | This study |
| YYS1918 | MATa Δ_hsm3_::KanMX [_YCplac22-HSM3pr-hsm3T190A-HA_5] | This study |
| YYS1919 | MATa Δ_hsm3_::KanMX [_YCplac22-HSM3pr-hsm3D230A-HA_5] | This study |
| YYS1920 | MATa Δ_hsm3_::KanMX [_YCplac22-HSM3pr-hsm3E157A/T190A-HA_5] | This study |
| YYS1921 | MATa Δ_hsm3_::KanMX [_YCplac22-HSM3pr-hsm3E157A/D230A-HA_5] | This study |
| YYS1922 | MATa Δ_hsm3_::KanMX [_YCplac22-HSM3pr-hsm3T190A/D230A-HA_5] | This study |
| YYS1923 | MATa Δ_hsm3_::KanMX [_YCplac22-HSM3pr-hsm3E157A/T190A/D230A-HA_5] | This study |
Native PAGE and Western Blotting
We analyzed proteasome assembly as described previously (19). In brief, yeast cells from an overnight culture in SC or SC-uracil media were diluted and shifted to 37 °C for 6 h, and then the cells (_A_600 of 1.5–1.8) were harvested and stored at −80 °C until use. The cells were lysed by glass beads, and 30 μg of cleared lysate was subjected to 3.5% native PAGE. After electrophoresis, in-gel peptidase activities against _N_-succinyl-LLVY-7-amido-4-methylcoumarin (Peptide Inc., Japan) were imaged using a LAS4000 IR (Fuji Film Inc., Japan). Western blotting was performed as described previously (19) with the following antibodies; anti-Rpt2 antibody (BioMol), anti-yeast 20 S proteasome antibody (38), and anti-HA antibody (16B12, Babco).
RESULTS AND DISCUSSION
Overall Structure of Hsm3
The structures of Hsm3 were determined in two different crystal forms. The first structure was determined in the orthorhombic crystal form, and the other was determined in the monoclinic crystal form. Both structures have two molecules in the asymmetric unit. The overall structures of these four molecules are similar (root mean square deviations (r.m.s.d.) range for 457 Cα atoms of 0.2–1.0 Å) (supplemental Fig. S1). We will use the A chain of the _P_212121 form for the discussion below.
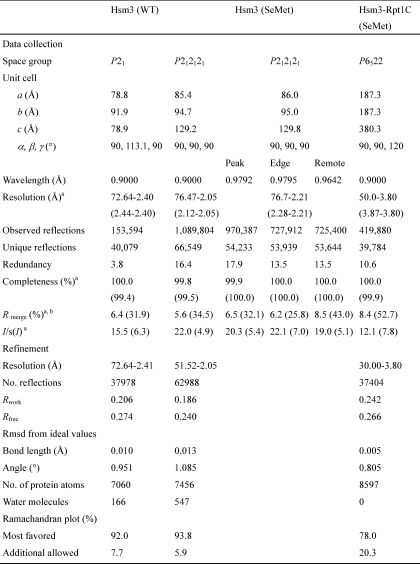
Hsm3 consists of 23 α-helices, α1–23 (Fig. 1A). The helices form 11 tandem HEAT repeats (HR1–11), each comprising two helices (referred as A and B) joined by a turn, with adjacent repeats connected by a linker. It has a C-shaped structure that consists entirely of helices and connecting loops. The diameter of Hsm3 is ∼86 Å; the central pore of the semicircle is 43 Å. The Hsm3 HEAT repeats are variable with the length of helices ranges from 22 to 7 residues, turns range from 12 to 3 residues, and linkers range from 10 to 2 residues (Fig. 1B). The fundamental architecture of the 11 HEAT repeats is similar, but the pairwise superposition of all repeats gives an r.m.s.d. ranging from 3.1 to 8.9 Å (supplemental Fig. S2). The A and B helices are not of equivalent length and are related by an inter-helix crossover angle of 10–70°. Comparison of the structures of Hsm3 with other known protein structures using the DALI server (39) revealed no marked structural similarities (Z score <13.8, r.m.s.d. = 9.3).
FIGURE 1.
Three-dimensional structure of yeast Hsm3. A, Two orthogonal views of the crystal structure of Hsm3. Each HEAT repeat of Hsm3 is labeled with HR1–HR11. The N and C termini of molecule are indicated. B, topological diagram of the secondary structure elements of Hsm3. The α-helices are represented by red circles (helix A in HR) and blue circles (helix B in HR), and the 310-helix is represented by yellow circles. Superscripts on the circles indicate the number of start and end residues of HEAT repeats. C, ribbon diagram of the two Hsm3 monomers in the asymmetric unit of the crystal. The N and C termini of each molecule are indicated.
The two molecules in the asymmetric unit of Hsm3 interact with each other, through their open sides of the C-shaped structure (Fig. 1C). The dimeric interactions are different between the two crystal forms. A total of 1700 Å2 of accessible surface area (850 Å2 for molecules A and B) is buried at the interface between Hsm3 dimer in _P_212121 form, whereas 1230 Å2 is buried in _P_21 form. This structural difference causes the distance between A and B molecules in the crystal. The distance between A and B molecules in _P_21 crystal is closer than that of _P_212121.
Structure of the Complex of Hsm3 with the C-terminal Domain of Rpt1
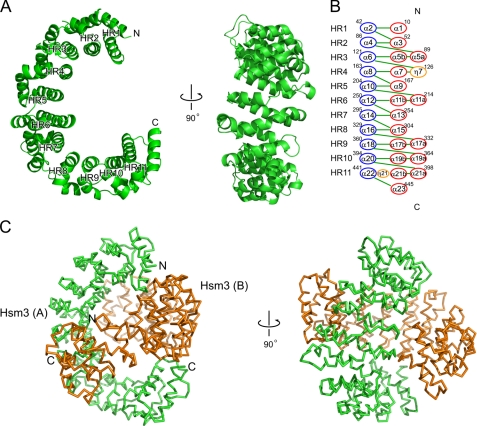
The previous study indicated that Hsm3 can directly interact with the Rpt1C (19). Hence, we determined the crystal structure of the Hsm3 complex with Rpt1C (Fig. 2A) in an attempt to understand the binding mode of Hsm3 to Rpt1. The electron density map at 3.8 Å resolution was combined with structural information obtained from the “methionine-marking” approach (supplemental Fig. S3) (40), allowing us to establish the structure of the Hsm3-Rpt1C complex. The asymmetric unit contains two heterodimers of Hsm3-Rpt1C (chains A-B and C-D). Overall structures of these two complexes are similar (r.m.s.d. range for 514 Cα atoms of 2.0 Å). As the complex involving chain A of Hsm3 and chain B of Rpt1C molecules is well defined, we will use it for the discussion below. The structure of Rpt1C (Fig. 2B), which consists of four α-helices (α1–4) that form an α-helical bundle, is essentially identical to the Rpt3C (supplemental Fig. S4); these structures have an r.m.s.d. of 1.6 Å for 53 Cα atoms (residues 396–449). Moreover, the C-terminal domain of Rpt1 can be superposed with those of the representative C-terminal domain of AAA+-ATPases PAN and HslU with an r.m.s.d. of 1.3 Å for 70 Cα atoms and 2.9 Å for 69 Cα atoms, respectively (Fig. 2C). The overall structure of Hsm3 in the Hsm3-Rpt1C complex is similar to that of the free Hsm3 structure with an average r.m.s.d. of 1.6 Å for 449 Cα positions, and the interacting regions for complex formation also superposed well. However, the diameter and the central pore of the semicircle of Hsm3 in the Hsm3-Rpt1C complex are changed (the diameter is ∼84 Å and the central pore of the semicircle is 38 Å) (Fig. 2D). This structural difference may influence changes in the conformation and orientation of the HR1 and HR11.
FIGURE 2.
Structure of the Hsm3 complexed with the C-terminal domain of Rpt1. A, overall architecture of the Hsm3-Rpt1C complex. Hsm3 (chain A) and Rpt1C (chain B) are colored green and orange, respectively. The N and C termini of each molecule are indicated. B, ribbon diagram of Rpt1C structure. Four α-helices are labeled from α1 to α4. C, structural comparison of C-terminal domains of three AAA+-ATPases as follows: Rpt1C (orange), PAN (PDB code 3H4M, cyan), and HslU (PDB code 1DO0, green). D, structural comparison between the free and the Rpt1C-bound Hsm3. The free Hsm3 (green) and the Hsm3 (red) in the complex with Rpt1C (orange) are shown.
Interaction between Hsm3 and Rpt1
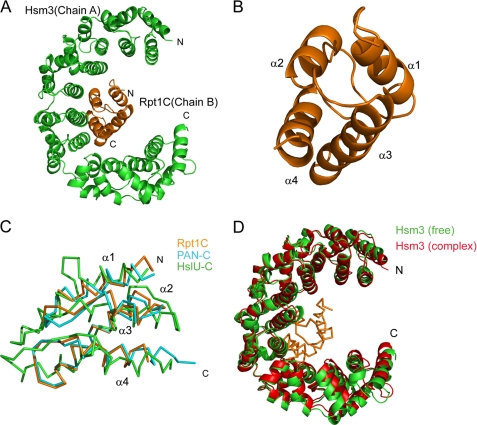
The interaction between Hsm3 and Rpt1C appeared to occur through α8, α10, and α12 of Hsm3 and helices α2, α4, and a loop α1–2 of Rpt1C by both hydrogen bonds and van der Waals contacts (Fig. 3A). A total of 953 Å2 of accessible surface area is buried at the interface between Hsm3 and Rpt1C. The Rpt1C binds to Hsm3 by packing its α2- and α4-helices against the concave surface of Hsm3. Residues involved in intermolecular formation are Glu-157, Thr-190, Asp-230, and Leu-232 of Hsm3 and Arg-390, Arg-403, Arg-409, Leu-410, and Leu-444 of Rpt1. Glu-157 and Thr-190 of Hsm3 make a potential salt bridge and hydrogen bond with Arg-409 and Arg-403, respectively. The interacting surfaces of these two proteins are hydrophobic core and complementary charged surface (Fig. 3B).
FIGURE 3.
Interface between Hsm3 and Rpt1. A, close-up view of the Hsm3-Rpt1C interface. A ribbon representation of the interacting regions of the complex. Some of the potential interacting residues are shown by stick representation. Hsm3 and Rpt1C are colored green and orange, respectively. B, surface potential representation of the Hsm3 and Rpt1C. The complementary surface patches responsible for complex formation are depicted by a circle. Red, blue, and white represent acidic, basic, and neutral, respectively.
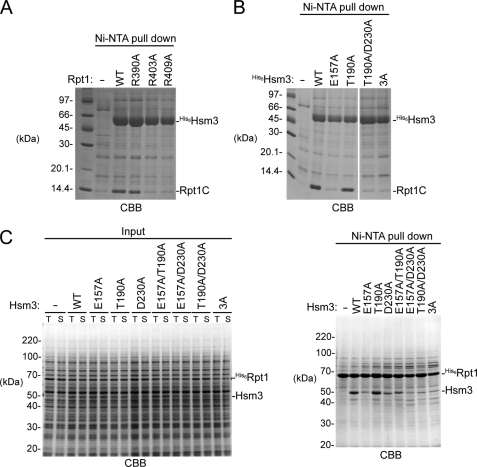
To investigate the roles of charged residues involved in the charged patches of Rpt1 and Hsm3, we performed binding assays in vitro (Fig. 4). Although the expression of the Rpt1C protein alone yielded an aggregated form in E. coli, Rpt1C was soluble when co-expressed with Hsm3, suggesting that Hsm3 has a potent chaperone activity to ensure Rpt1C solubility. We next mutated the charged resides of the Hsm3-Rpt1C interface to Ala (R390A, R403A, and R409A for Rpt1C and E157A, T190A, and D230A in Hsm3) and carried out a Ni-NTA pulldown experiment to monitor the complex formation. The R403A and R409A mutation of Rpt1C reduced the binding ability to Hsm3, whereas the R390A mutant had little effect (Fig. 4A). This result suggested that Arg-403 and Arg-409 in Rpt1C are critical residues for Hsm3 binding. However, the E157A mutation in Hsm3 reduced the Rpt1C binding, whereas the T190A mutant had no effect (Fig. 4B). The D230A and the E157A/D230A mutants were not expressed in E. coli probably due to protein folding problem. Therefore, we further investigated the interaction using a wheat germ cell-free system, which has the advantage of producing difficult-to-express proteins in E. coli (33, 34). Although bacterially expressed His6-tagged full-length Rpt1 was usually detected in insoluble fraction (data not shown), almost all the His6-Rpt1 protein was found in a soluble fraction by using the cell-free system even if Hsm3 protein was not co-synthesized (Fig. 4C, left panel). As observed in the case of co-expression in E. coli, the E157A mutation in Hsm3 greatly reduced the Rpt1 binding (Fig. 4C, right panel). Furthermore, the D230A mutation in Hsm3 markedly decreased the Hsm3-Rpt1. Thus, two acidic residues, Glu-157 and Asp-230, of Hsm3 contribute to the Rpt1 recognition.
FIGURE 4.
Characterization of the crucial residues in the Hsm3-Rpt1 interface. A, pulldown assay from bacterially co-expressing His6-Hsm3 and mutant versions of Rpt1C. Eluted proteins from Ni-NTA resin were subjected to SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining. B, pulldown assay from bacterially co-expressing His6-Hsm3 mutants and Rpt1C. Eluted proteins from Ni-NTA resin were subjected to SDS-PAGE followed by Coomassie Brilliant Blue staining. Triple mutation of Hsm3 (E157A/T190A/D230A) is indicated in Fig. 3A. C, pulldown assay between full-length His6-Rpt1 and Hsm3 mutants. His6-fused full-length Rpt1 and indicated Hsm3 mutants were co-synthesized by a wheat germ cell-free expression system and then pulled down by Ni-NTA resin. Input and eluted proteins were subjected to SDS-PAGE followed by Coomassie Brilliant Blue staining. Total reaction and supernatant are labeled as T and S, respectively.
Mutations of the Hsm3-Rpt1 Interface Cause the Assembly Defect of the 26 S Proteasome in Vivo
To determine the physiological relevance of the Hsm3-Rpt1 interaction, we generated a series of yeast mutant strains that harbor the mutations in the Hsm3-Rpt1 interface and analyzed the proteasome assembly by native-PAGE (Fig. 5). Because of the inability of efficient formation of the Rpt1-Rpt2-Rpn1 complex, the 26 S proteasome levels were moderately reduced in the Hsm3 knock-out cells (16–19). If the charged residues were critical for the Hsm3-Rpt1 interactions, the mutants should be a phenocopy of the Δ_hsm3_.
FIGURE 5.
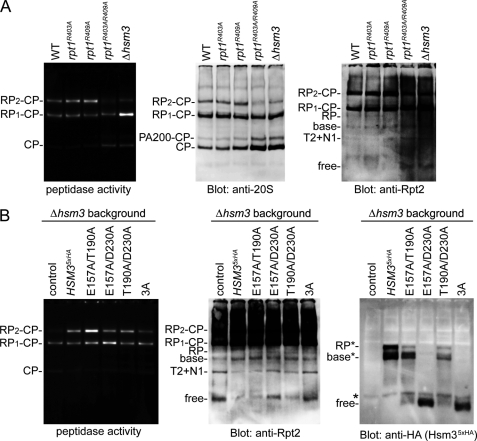
Mutations of the Hsm3-Rpt1 interface cause assembly defect of the 26 S proteasome. A, native-PAGE analysis of the 26 S proteasome assembly in rpt1 mutants. Extracts of the indicated mutants were subjected to native-PAGE. After electrophoresis, peptidase activities of the proteasome were visualized by in gel peptidase assay (left). The same gel was analyzed by Western blotting with the indicated antibodies. RP_2_-CP and RP_1_-CP indicate doubly and singly capped 26 S proteasomes, respectively. Rpn1 and Rpt2 are abbreviated as N1 and T2, respectively. B, native-PAGE analysis of the 26 S proteasome assembly in hsm3 mutants. HA-tagged Hsm3 and its mutants were expressed under the native promoter in the Δ_hsm3_ cells. Extracts from the indicated mutants were analyzed as in A. RP* and base* seemed to be transient complexes that contain Hsm3. Hsm3 mutants that harbor triple mutations of Hsm3 (E157A/T190A/D230A) are indicated as 3A.
We first analyzed the effects of the rpt1 mutations on the proteasome assembly. Although in vitro binding assays suggested a single mutation of Arg-403 or Arg-409 in Rpt1 caused the loss of Hsm3 binding (Fig. 4), the assembly defect of the proteasome was not observed in the rpt1R403A and the rpt1R409A single mutant cells. However, in the rpt1R403A/R409A mutant, the 26 S proteasome levels were markedly reduced, whereas the free 20 S CP and the free Rpt2 were increased (Fig. 5A). The result suggested both residues of Rpt1 are responsible for the Hsm3 binding and the proteasome assembly.
Similarly, although in vitro binding assays suggested that Glu-157 of Hsm3 is a critical residue for the Rpt1 recognition (Fig. 4), no significant assembly defect was detected in the hsm3E157A mutant cells (data not shown). However, we detected assembly defects in the hsm3 mutants with multiple mutations, especially in the hsm3E157A/D230A and the hsm3E157A/T190A/D230A cells (Fig. 5B), suggesting that E157 and D230 are important residues for the Rpt1 binding. Collectively, these results indicate that the complementary charged interactions, the Rpt1R409-Hsm3E157 and the Rpt1R403-Hsm3D230, are a critical role for the RP assembly.
It should be noted that neither the rpt1R403A/R409A mutant nor the hsm3E157A/T190A/D230A mutant cells showed sensitivity to high temperature as seen in the Δ_hsm3_ mutant cells (16–19). Remaining weak interactions in the mutants, probably due to the hydrophobic interactions, may be sufficient for the Hsm3-Rpt1 interaction, although the 26 S proteasome levels are considerably reduced.
Structural Features and Functional Similarity among Base Assembly Chaperones
The tertiary structures of Hsm3, Rpn14, and Nas6 are quite different; Hsm3 contains HEAT repeats, Rpn14 contains WD40 motifs, and Nas6 contains ankyrin repeats. Nevertheless, these chaperones bind the C-terminal regions of specific Rpt subunits, respectively. Nas6 was shown to interact specifically with the C-terminal region of Rpt3 through surface charge complementarities, i.e. one positively and two negatively charged patches in Nas6 and one negatively and two positively charged patches in Rpt3C (supplemental Fig. S5_A_ and B) (40). The interacting surface Rpn14 was also predicted, i.e. negatively charged patches in Rpn14 and positively charged patches in Rpt6 (20).
To understand their molecular basis for binding specificity, we compared the structures of interfaces between Rpt subunits and chaperones. The C-terminal domains of Rpt1 and Rpt3 showed that the overall tertiary structures are similar, and the interacting regions of each Rpt subunit for complex formation are similar (supplemental Fig. S5_C_). However the charged patches on the interface are significantly different between Hsm3 and Nas6. Whereas α2, α4, and the loops α1–2, α2–3, and α3–4 of Rpt3C make contact with Nas6, α2, α4, and loop α1–2 of Rpt1C interact with Hsm3. Most of the interacting residues responsible for making the complexes are not conserved between Rpt1 and Rpt3 (supplemental Fig. S6). We assumed that these differences account for the specificities of Rpt subunits by the 19 S base-dedicated chaperones.
Structural Implication of Molecular Mechanisms of Hsm3 for Base Assembly
The structure of the Hsm3-Rpt1C complex illustrates an intermediate state of proteasome assembly. A model of Hsm3 interacting with the ATPase domain of PAN (residues 158–418) and the AAA-ATPase ring were generated by superimposing the C-terminal domain of Rpt1 from Hsm3-Rpt1C on the structure of the PAN and HslU (Fig. 6A). The C-terminal domain of Rpt1 shows 39 and 9% sequence identities with PAN and HslU (r.m.s.d. range for Cα atoms of 1.3 and 2.9 Å), respectively. In the Hsm3-PAN complex model, the C-terminal region of Hsm3 causes steric hindrance with Rpt1 (supplemental Fig. S7). However, the full-length Rpt1 can bind Hsm3 (Fig. 4C) suggesting that the C-terminal region of Hsm3 might accommodate structural changes upon binding to Rpt1. In the model, Hsm3 and ATPase domain of PAN are bound by interactions of loop α21–α22 and helix α23 of Hsm3 and loop β3–α5 and loop α6–α7 of the PAN ATPase domain. The residues involved in the Hsm3 interactions of the β3–α5 loop of PAN are not conserved among Rpt subunits, whereas those participating in the α6–α7 loop are conserved. The C-terminal region of Hsm3 may also participate in the molecular recognition of β3–α5 loop of Rpt1 ATPase domain.
FIGURE 6.
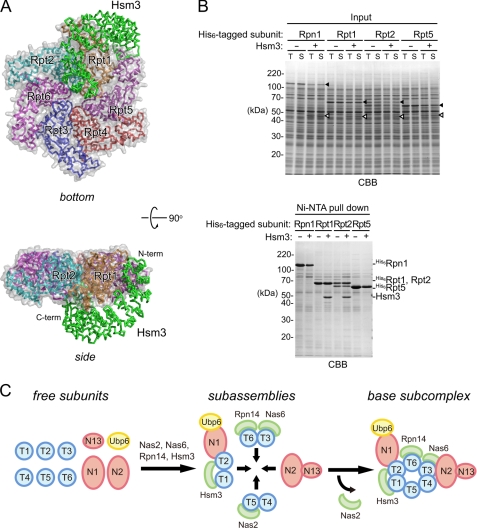
Hsm3 is a scaffold protein for Rpt1-Rpt2 formation. A, structural model of Hsm3-Rpt ring complex. The Hsm3-Rpt1C structure was superimposed on a structural model of hexameric ATPase (ATPase ring model was generated from the HslU (PDB code 1DO0) structure). Hsm3 and Rpt ring are shown as Cα trace representations and are colored green (Hsm3), orange (Rpt1), cyan (Rpt2), blue (Rpt3), red (Rpt4), purple (Rpt5), and pink (Rpt6), respectively. Surfaces of Rpt subunits are shown as surface plots (gray). B, in vitro pulldown assay between Hsm3 and His6-tagged base subunits. His6-tagged full-length Rpt1, Rpt2, Rpt5, or Rpn1 were synthesized with or without Hsm3 by a wheat germ cell-free expression system. Interactions between the His6-tagged base subunits and Hsm3 were analyzed by pulldown assay with Ni-NTA resin. Input and eluted proteins were subjected to SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining. The protein bands corresponding to the base subunits and Hsm3 are indicated by filled and open arrowheads, respectively. C, model of the base assembly. Four assembly intermediates are formed from nine free base subunits and a deubiquitylating enzyme Ubp6 (42). This study suggests that Hsm3 plays a scaffolding role in the formation of the Rpt1 and Rpt2 complex. Finally, the four intermediates are jointed into the base subcomplex. Nas2 is dissociated upon binding of Nas2 module and Nas6 module (43). Rpn and Rpt are abbreviated as N and T, respectively.
In the Hsm3-ATPase ring model, HR10 and HR11 of Hsm3 create steric clashes with the loop α6–α7 region of Rpt2. However, a major Hsm3 containing assembly intermediate is Hsm3-Rpt1-Rpt2-Rpn1 in the cells (16–19). Instead, the model raised the possibility that Hsm3 may associate with other neighboring subunits in addition to Rpt1. Using the wheat germ cell-free system, we investigated whether Hsm3 can interact with Rpn1, Rpt2, and Rpt5 (Fig. 6B). Strikingly, we observed a direct interaction between Hsm3 and Rpt2, consistent with a previous study of mammalian proteasomes, in which Gorbea et al. (41) showed that S5b interacts with the AAA-ATPase domain of Rpt2. Thus, these results indicate that Hsm3/S5b has a scaffolding role to assist the interaction between Rpt1 and Rpt2 (Fig. 6C). According to our results and recent studies (42, 43), we update a model for the base assembly (Fig. 6C). Nine base subunits and a deubiquitylating enzyme Ubp6 are assembled into four distinct base intermediates (Hsm3 module, Hsm3-Rpt1-Rpt2-Rpn1-Ubp6; Nas2 module, Nas2-Rpt5-Rpt4; Nas6 module, Nas6-Rpt3-Rpt6-Rpn14; and Rpn2-Rpn13). Three of four intermediates are assisted by the RP-dedicated chaperones, Nas2, Nas6, Rpn14, and Hsm3. Hsm3 plays a scaffolding role in the formation of the Rpt1 and Rpt2 complex. Nas2 is dissociated upon binding of the Nas2 module and Nas6 module (43), and subsequently, the remaining subassemblies are incorporated and form the base subcomplex of the RP.
In summary, our results provided structural insights in the molecular mechanisms of the recognition of Rpt1 and also how Hsm3 assists the molecular assembly of the RP of the 26 S proteasome.
Supplementary Material
Supplemental Data
Acknowledgments
We thank all members of beamline BL44XU for help in data collection at SPring-8.
*
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, grant-in-aid for scientific research on innovative areas (to T. M.), grant-in-aid for challenging exploratory research (to T. M.), The Naito Foundation (to T. M.), grant-in-aid for JSPS fellows (to K. Takagi), grant-in-aid for young scientists (to Y. S.), grant-in-aid for specially promoted Research (to K. Tanaka), and the Targeted Proteins Research program (to R. M., Y. E., K. K., K. Tanaka, and T. M.).
♦
This article was selected a Paper of the Week.
The atomic coordinates and structure factors (codes 3VLD, 3VLE, and 3VLF) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
4
The abbreviations used are:
CP
core particle
RP
regulatory particle
HEAT
huntingtin-elongation-A subunit-TOR
PDB
Protein Data Bank
Ni-NTA
nickel nitrilotriacetic acid
r.m.s.d.
root mean square deviation
SeMet
selenomethionine.
REFERENCES
- 1.Baumeister W., Walz J., Zühl F., Seemüller E. (1998) The proteasome. Paradigm of a self-compartmentalizing protease. Cell 92, 367–380 [DOI] [PubMed] [Google Scholar]
- 2.Coux O., Tanaka K., Goldberg A. L. (1996) Structure and functions of the 20 S and 26 S proteasomes. Annu. Rev. Biochem. 65, 801–847 [DOI] [PubMed] [Google Scholar]
- 3.Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 [DOI] [PubMed] [Google Scholar]
- 5.Leggett D. S., Glickman M. H., Finley D. (2005) Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 [DOI] [PubMed] [Google Scholar]
- 6.Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. (2010) Assembly, structure, and function of the 26 S proteasome. Trends Cell Biol. 20, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murata S., Yashiroda H., Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 8.Tomko R. J., Jr., Hochstrasser M. (2011) Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell. Biochem. Biophys. 60, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano Y., Kaneko T., Okamoto K., Bai M., Yashiroda H., Furuyama K., Kato K., Tanaka K., Murata S. (2008) Dissecting β-ring assembly pathway of the mammalian 20 S proteasome. EMBO J. 27, 2204–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos P. C., Höckendorff J., Johnson E. S., Varshavsky A., Dohmen R. J. (1998) Ump1p is required for proper maturation of the 20 S proteasome and becomes its substrate upon completion of the assembly. Cell 92, 489–499 [DOI] [PubMed] [Google Scholar]
- 11.Hirano Y., Hendil K. B., Yashiroda H., Iemura S., Nagane R., Hioki Y., Natsume T., Tanaka K., Murata S. (2005) A heterodimeric complex that promotes the assembly of mammalian 20 S proteasomes. Nature 437, 1381–1385 [DOI] [PubMed] [Google Scholar]
- 12.Le Tallec B., Barrault M. B., Courbeyrette R., Guérois R., Marsolier-Kergoat M. C., Peyroche A. (2007) 20 S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell 27, 660–674 [DOI] [PubMed] [Google Scholar]
- 13.Hirano Y., Hayashi H., Iemura S., Hendil K. B., Niwa S., Kishimoto T., Kasahara M., Natsume T., Tanaka K., Murata S. (2006) Cooperation of multiple chaperones required for the assembly of mammalian 20 S proteasomes. Mol. Cell 24, 977–984 [DOI] [PubMed] [Google Scholar]
- 14.Kusmierczyk A. R., Kunjappu M. J., Funakoshi M., Hochstrasser M. (2008) A multimeric assembly factor controls the formation of alternative 20 S proteasomes. Nat. Struct. Mol. Biol. 15, 237–244 [DOI] [PubMed] [Google Scholar]
- 15.Yashiroda H., Mizushima T., Okamoto K., Kameyama T., Hayashi H., Kishimoto T., Niwa S., Kasahara M., Kurimoto E., Sakata E., Takagi K., Suzuki A., Hirano Y., Murata S., Kato K., Yamane T., Tanaka K. (2008) Crystal structure of a chaperone complex that contributes to the assembly of yeast 20 S proteasomes. Nat. Struct. Mol. Biol. 15, 228–236 [DOI] [PubMed] [Google Scholar]
- 16.Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Tallec B., Barrault M. B., Guérois R., Carré T., Peyroche A. (2009) Hsm3/S5b participates in the assembly pathway of the 19 S regulatory particle of the proteasome. Mol. Cell 33, 389–399 [DOI] [PubMed] [Google Scholar]
- 18.Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saeki Y., Toh-e A., Kudo T., Kawamura H., Tanaka K. (2009) Multiple proteasome-interacting proteins assist the assembly of the yeast 19 S regulatory particle. Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 20.Kim S., Saeki Y., Fukunaga K., Suzuki A., Takagi K., Yamane T., Tanaka K., Mizushima T., Kato K. (2010) Crystal structure of yeast rpn14, a chaperone of the 19 S regulatory particle of the proteasome. J. Biol. Chem. 285, 15159–15166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22.Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z. (1991) Proceedings of the CCP4 Study Weekend (Wolf W., Evans P. R., Leslie A. G. W., eds) pp. 80–86, Warrington, Daresbury Laboratory [Google Scholar]
- 24.Cowtan K. (1999) Error estimation and bias correction in phase-improvement calculations. Acta Crystallogr. D Biol. Crystallogr. 55, 1555–1567 [DOI] [PubMed] [Google Scholar]
- 25.Morris R. J., Perrakis A., Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28.Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 29.Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 30.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 31.DeLano W. (2002) PyMOL, De Lano Scientific, San Carlos, CA [Google Scholar]
- 32.McNicholas S., Potterton E., Wilson K. S., Noble M. E. (2011) Presenting your structures. The CCP4mg molecular graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goshima N., Kawamura Y., Fukumoto A., Miura A., Honma R., Satoh R., Wakamatsu A., Yamamoto J., Kimura K., Nishikawa T., Andoh T., Iida Y., Ishikawa K., Ito E., Kagawa N., Kaminaga C., Kanehori K., Kawakami B., Kenmochi K., Kimura R., Kobayashi M., Kuroita T., Kuwayama H., Maruyama Y., Matsuo K., Minami K., Mitsubori M., Mori M., Morishita R., Murase A., Nishikawa A., Nishikawa S., Okamoto T., Sakagami N., Sakamoto Y., Sasaki Y., Seki T., Sono S., Sugiyama A., Sumiya T., Takayama T., Takayama Y., Takeda H., Togashi T., Yahata K., Yamada H., Yanagisawa Y., Endo Y., Imamoto F., Kisu Y., Tanaka S., Isogai T., Imai J., Watanabe S., Nomura N. (2008) Human protein factory for converting the transcriptome into an _in vitro_-expressed proteome,. Nat. Methods 5, 1011–1017 [DOI] [PubMed] [Google Scholar]
- 34.Takai K., Sawasaki T., Endo Y. (2010) Practical cell-free protein synthesis system using purified wheat embryos. Nat. Protoc. 5, 227–238 [DOI] [PubMed] [Google Scholar]
- 35.Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 36.Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki T., Toh-e A., Kikuchi Y. (2000) Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40 S ribosome biogenesis in the nucleolus. Mol. Cell Biol. 20, 7971–7979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka K., Yoshimura T., Kumatori A., Ichihara A., Ikai A., Nishigai M., Kameyama K., Takagi T. (1988) Proteasomes (multiprotease complexes) as 20 S ring-shaped particles in a variety of eukaryotic cells. J. Biol. Chem. 263, 16209–16217 [PubMed] [Google Scholar]
- 39.Holm L., Park J. (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 40.Nakamura Y., Umehara T., Tanaka A., Horikoshi M., Padmanabhan B., Yokoyama S. (2007) Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26 S proteasome. Biochem. Biophys. Res. Commun. 359, 503–509 [DOI] [PubMed] [Google Scholar]
- 41.Gorbea C., Taillandier D., Rechsteiner M. (2000) Mapping subunit contacts in the regulatory complex of the 26 S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J. Biol. Chem. 275, 875–882 [DOI] [PubMed] [Google Scholar]
- 42.Sakata E., Stengel F., Fukunaga K., Zhou M., Saeki Y., Förster F., Baumeister W., Tanaka K., Robinson C. V. (2011) The catalytic activity of Ubp6 enhances maturation of the proteasomal regulatory particle. Mol. Cell 42, 637–649 [DOI] [PubMed] [Google Scholar]
- 43.Tomko R. J., Jr., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010) Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases. Implications for proteasome structure and assembly. Mol. Cell 38, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data