Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model (original) (raw)
Abstract
The reconstruction of the Tree of Life has relied almost entirely on concatenation methods, which do not accommodate gene tree heterogeneity, a property that simulations and theory have identified as a likely cause of incongruent phylogenies. However, this incongruence has not yet been demonstrated in empirical studies. Several key relationships among eutherian mammals remain controversial and conflicting among previous studies, including the root of eutherian tree and the relationships within Euarchontoglires and Laurasiatheria. Both Bayesian and maximum-likelihood analysis of genome-wide data of 447 nuclear genes from 37 species show that concatenation methods indeed yield strong incongruence in the phylogeny of eutherian mammals, as revealed by subsampling analyses of loci and taxa, which produced strongly conflicting topologies. In contrast, the coalescent methods, which accommodate gene tree heterogeneity, yield a phylogeny that is robust to variable gene and taxon sampling and is congruent with geographic data. The data also demonstrate that incomplete lineage sorting, a major source of gene tree heterogeneity, is relevant to deep-level phylogenies, such as those among eutherian mammals. Our results firmly place the eutherian root between Atlantogenata and Boreoeutheria and support ungulate polyphyly and a sister-group relationship between Scandentia and Primates. This study demonstrates that the incongruence introduced by concatenation methods is a major cause of long-standing uncertainty in the phylogeny of eutherian mammals, and the same may apply to other clades. Our analyses suggest that such incongruence can be resolved using phylogenomic data and coalescent methods that deal explicitly with gene tree heterogeneity.
Keywords: gene tree heterogeneity, incomplete lineage sorting, multispecies coalescent model, phylogenetic incongruence
To date, phylogenetic studies using DNA sequence data have been based almost entirely on concatenation methods. Concatenation methods infer phylogenies from multilocus sequences that are combined to form a single supermatrix (1), based on the assumption that all genes have the same or similar phylogenies (1, 2). However, empirical studies have shown widespread presence of gene tree heterogeneity within mammals and other clades (3, 4). When a high level of gene tree heterogeneity occurs in multilocus sequence data, theory and simulations have predicted that concatenation methods can yield misleading results (5, 6). By contrast, more recently developed coalescence-based methods estimate a species phylogeny from a collection of gene trees, an approach that allows different genes to have different topologies (4, 7–10). Simulations and theory have shown that coalescent methods can produce accurate phylogenies from multilocus sequence data that are subject to incomplete lineage sorting (ILS), a major cause of gene tree heterogeneity (4, 7–10). However, the superior performance of coalescent methods relative to concatenation methods in the face of substantial gene tree heterogeneity remains to be demonstrated in empirical studies.
Resolving the phylogeny of eutherian mammals has been challenging due to conflicting results from previous studies (11–20). In the past decade, the division of eutherian mammals into four superorders—Euarchontoglires, Laurasiatheria, Afrotheria, and Xenarthra—has been well supported (11–20). However, some key elements of eutherian mammal relationships, including the root of the eutherian tree and the interordinal relationships within Euarchontoglires and Laurasiatheria, remain unresolved or unstable (20). Resolving these incongruences is crucial not only for understanding the evolutionary history and dynamics of Eutheria, but also for revealing the source of contradictions on eutherian phylogeny in previous studies. Using a phylogenetic, DNA-based analysis of eutherian mammal relationships as a case study, we empirically demonstrate that concatenation methods can lead to phylogenetic results that are inherently incongruent, in that different subsamples of the same data set tend to produce strongly divergent topologies. Analyzing and subsampling the same data using coalescent methods yield more consistent results, and the resulting phylogeny suggests possible resolutions to persistent controversies regarding the position of the root of Eutheria and key relationships within Laurasiatheria and Euarchontoglires.
Results
Conflict Between Concatenation and Coalescent Phylogenetic Analyses.
We analyzed sequence data from 447 nuclear genes from 33 eutherian species representing 16 of 18 eutherian orders and four outgroups including two marsupials, one monotreme, and chicken. The 447 orthologous genes in the data are distributed across all 22 autosomes and the X chromosome in the human genome, allowing us to access the phylogenetic utility of different parts across the genome.
Our analyses used two recently developed coalescent methods: the Maximum Pseudolikelihood Estimation of the Species Tree (MP-EST) method (8) and the Species Tree Estimation using Average Ranks of coalescence (STAR) method, used here with the neighbor-joining algorithm (9). MP-EST uses the frequencies of gene trees of triplets of taxa to estimate the topology and branch lengths (in coalescent units) of the overall species tree (8), whereas STAR computes the topological distances among pairs of taxa as the average of the ranks (number of nodes toward the root node) of those taxon pairs across nodes in the collected gene trees (9). MP-EST and STAR are partially parametric methods that reconstruct species phylogenies using only the topology of gene trees based on summary statistics, whereas fully parametric methods use all aspects of the data to infer phylogenies (4, 21). Because partially parametric methods use only part of the information contained in the data, they usually require more loci than fully parametric methods to achieve a certain level of confidence in the results (4, 21). However, partially parametric methods have computational advantages because these methods can quickly infer phylogenies from large-scale genomic data. In contrast, it is difficult to apply fully parametric methods to such data sets due to their extensive computational demands. Additionally, MP-EST and STAR are robust to violation of the assumptions that underpin many coalescent analyses. Because both methods are based on summary statistics calculated across all gene trees, a small number of outlier genes that significantly deviate from the coalescent model have little effect on the ability of either method to accurately reconstruct species trees. We compared the results from both coalescent methods with those from concatenation analyses implemented in two popular phylogenetic algorithms, MrBayes (Bayesian) (22) and RAxML (maximum likelihood) (23).
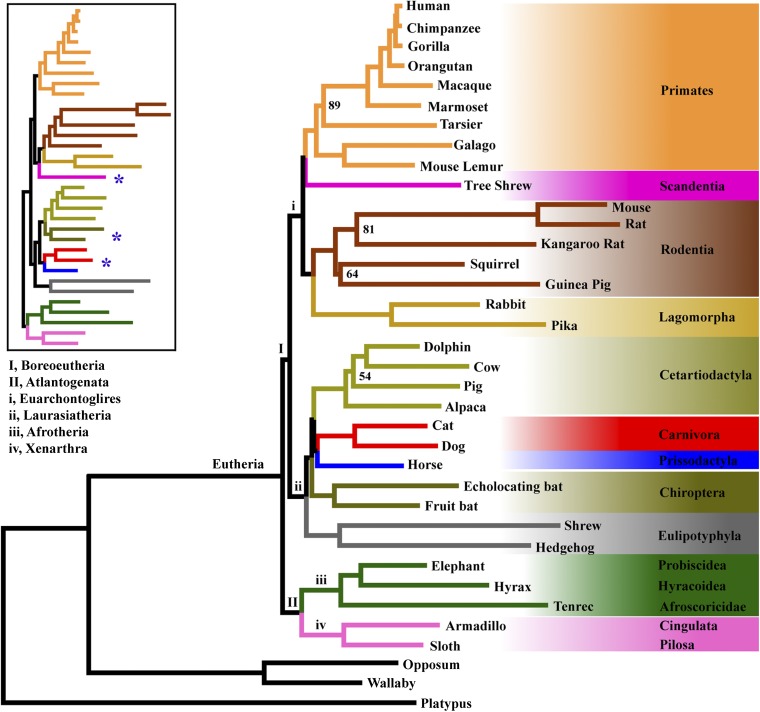
Trees obtained by coalescent analyses of the full data set consistently support the following evolutionary relationships among eutherian mammals (Fig. 1 and SI Appendix, Fig. S1): Afrotheria and Xenarthra form a strongly supported monophyletic clade Atlantogenata [Bootstrap percentage (BP) = 100% by both MP-EST and STAR], which comprises the sister taxon of Boreoeutheria; within Euarchontoglires, Scandentia (tree shrews) constitutes the sister group of primates (BP = 99% by MP-EST, 94% by STAR); within Laurasiatheria, Perissodactyla and Carnivora form a monophyletic group (BP = 96% by MP-EST, 98% by STAR) that is sister to Cetartiodactyla (BP = 90% by MP-EST, 94% by STAR); Chiroptera is the sister group of the clade comprising Cetartiodactyla, Perissodactyla, and Carnivora (BP = 99% by MP-EST, 100% by STAR); and Eulipotyphla forms the basal branch of Laurasiatheria (BP = 100% by both MP-EST and STAR).
Fig. 1.
Evolutionary relationships of eutherian mammals. The phylogeny was estimated using the maximum-pseudolikelihood coalescent method MP-EST with multilocus bootstrapping (8, 40). The numbers on the tree indicate bootstrap support values, and nodes with bootstrap support >90% are not shown. Branch lengths were estimated by fitting the concatenated sequence data for all 447 loci to the MP-EST topology using standard ML and an appropriate substitution model in PAUP* v.4.0 (45). (Inset) The eutherian phylogeny estimated using the Bayesian concatenation method implemented in MrBayes (22). The ML concatenation tree built by RAxML (23) is identical to the Bayesian concatenation tree in topology. Branches of the concatenation tree are coded by the same colors as in the MP-EST tree. The blue asterisks indicate the position of Scandentia (tree shrews), Chiroptera (bats), Perissodactyla (odd-toed ungulates), and Carnivora (carnivores), whose placement differs from the coalescent tree. The Bayesian concatenation tree received a posterior probability support of 1.0 for all nodes. In SI Appendix, Fig. S2, the concatenation tree with taxon names is shown.
The trees made by concatenation methods are similar to the coalescent trees, with the following key differences (Fig. 1; SI Appendix, Figs. S2 and S3): within Euarchontoglires, Scandentia is the sister group of Glires rather than Primates; and within Laurasiatheria, Chiroptera and Cetartiodactyla constitute a monophyletic group that is sister to the clade formed by Perissodactyla and Carnivora. The concatenation trees received a posterior probability of 1.0 or bootstrap support >90% for all nodes except the group of Chiroptera and Cetartiodactyla with BP = 80% (SI Appendix, Fig. S3).
Source of Phylogenetic Conflict Revealed by Subsampling of Loci and Taxa.
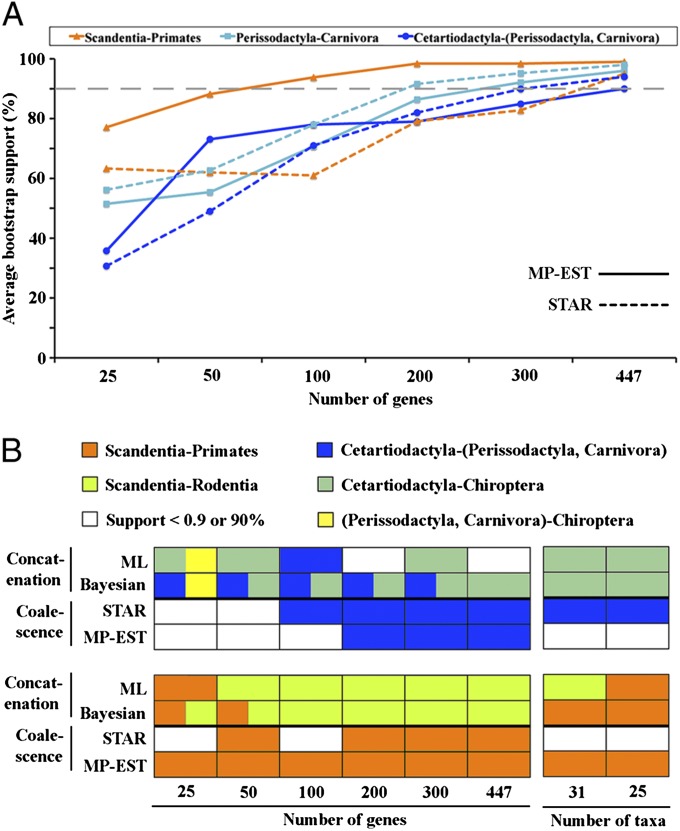
To resolve the incongruence in the results, we evaluated the effect of subsampling of loci and taxa on the performance of phylogenetic methods. We constructed coalescent and concatenation trees for different gene sets that include 25, 50, 100, 200, and 300 genes, randomly selected from the 447-gene set with 10 replicates for each gene set. It is an expectation of phylogenetic methodology that nodal support values should increase with increasing number of loci and that the highly supported clades remain the same rather than changing erratically as more data are collected. Consistent with this prediction, the coalescent analyses estimate a consistent phylogeny for eutherian mammals using different subsets of loci and show a clear trend of increasing support for weakly resolved nodes with increasing numbers of loci (Fig. 2_A_). In contrast, concatenation analyses assigned high support (PP > 0.9 or BP > 90%) for conflicting relationships among eutherian mammals based on different subsets of loci (Fig. 2_B_). For example, the interordinal relationships within Laurasiatheria vary across concatenation trees estimated from different data sets; however, all these different relationships received high support or complete support (Fig. 2_B_). The high support for incongruent relationships suggests that concatenation methods have misled the node support values. Excessive posterior probabilities were recognized early on in phylogenetic analyses using Bayesian concatenation (12, 24), although the phylogenetically erratic behavior of concatenation analyses with different data sets has been previously unrecognized in empirical studies.
Fig. 2.
Trends in bootstrap support for coalescent analyses and incongruence of concatenation estimates for eutherian phylogeny. (A) Gradual increase in bootstrap support values with increasing gene numbers using coalescent methods for three clades: Scandentia–Primates within Euarchontoglires, Perissodactyla–Carnivora and Cetartiodactyla–(Perissodactyla, Carnivora) within Laurasiatheria. The gray dashed line indicates bootstrap support of 90%. (B) Concatenation analyses yield conflicting phylogenies within Euarchontoglires and Laurasiatheria for subsampled gene and taxon sets. We constructed coalescent and concatenation trees for different sets of 25, 50, 100, 200, and 300 genes randomly selected from the 447-gene set, with 10 replicates for each gene set except 447. We also constructed trees for two reduced taxon sets by excluding 6 and 12 eutherian taxa. White cells in the heatmap indicate that the support for all replicates is <0.9 or 90%. Colored cells indicate relationships that received node support values >0.9 or 90% for at least one replicate. Cells with two colors indicate two highly supported but conflicting relationships among different replicates. Note that the concatenation analyses frequently support conflicting relationships for different gene and taxon sets, whereas the coalescent methods consistently support the same topology.
We also tested the influence of taxon sampling on the performance of phylogenetic methods by excluding 6 and 12 eutherian taxa from the original data set and repeating the phylogenetic analyses. Coalescent analyses again gave a consistent phylogenetic estimate of relationships, whereas both Bayesian and ML concatenation methods yielded misleading phylogenies with excessive nodal support values (Fig. 2_B_; SI Appendix, Figs. S5–S8). The sensitivity to variable taxon sampling therefore constitutes an additional challenge for concatenation methods.
Relevance of Incomplete Lineage Sorting for Deep-Level Clades.
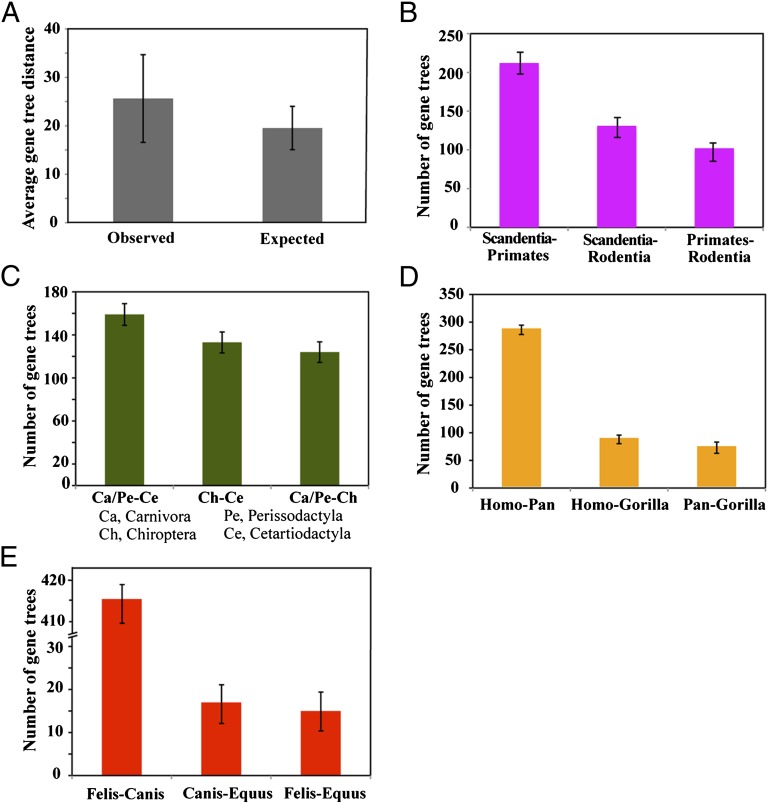
It has been widely assumed that ILS is relevant only to recent radiations as a source of gene tree heterogeneity (25, 26). To test this, we reconstructed individual gene trees from each of the 447 loci using maximum likelihood (ML) (23) and measured the extent of gene tree variation in topology as well as the distribution of gene tree relationships across particular clades. Overall we found 440 topologically distinct trees in the full data set, indicating that the tree for nearly every gene is distinct. The low consensus values for nodes of the consensus of gene trees also indicate a substantial level of gene tree heterogeneity (SI Appendix, Fig. S4). Nonetheless, through simulations, we estimate that the multispecies coalescent model accounts for 77% of the variation in gene trees in the full data set (Fig. 3_A_). When gene tree heterogeneity is caused by ILS, the multispecies coalescent model predicts that, for nodes of triplets of species, the two minority triplet gene trees should be equally frequent (10, 27). Consistent with this prediction, the frequencies of minority gene trees are similar for nodes where gene tree heterogeneity is present (Fig. 3 B_–_E). These analyses suggest that ILS is relevant even to deep-level clades of eutherian mammals. This result is expected, even though it was previously difficult to demonstrate due to potentially low phylogenetic signal, because theory suggests that it is only the length of internodes as measured in coalescent units, not the relative or absolute depth of those internodes in a given tree, that is relevant for the presence of ILS (28).
Fig. 3.
The mammal data set is consistent with the multispecies coalescent model. (A) Distribution of expected and observed gene tree distances. Expected gene trees were simulated from the MP-EST species tree under the multispecies coalescent model. Observed gene trees were estimated from the 447 genes in the full data set. Gene tree distances were calculated using standard measures (27). Note that the expected gene tree distance can account for about 77% of the observed gene tree distance. (B_–_E) Distribution of majority and minority gene tree triplets for specific eutherian clades. In cases where one of the three taxa in the triplet consists of multiple species, we counted the frequency of all relevant gene tree triplets for a given gene and then assigned the majority triplet to that gene. Ties were ignored, and hence the totals sometimes do not sum to 447 genes.
Our data set is also noteworthy in using loci that are relatively long compared with individual loci used in traditional phylogenetic studies. For example, the average length of loci in our data set is ∼3.1 kb (1 SD = 2,334), with seven loci greater than 10 kb (SI Appendix, Fig. S9). However, long loci have the disadvantage of being more susceptible to recombination within loci, which could have occurred within species or in the common ancestors in our tree. This would constitute a violation of the multispecies coalescent model and is one factor known to mislead phylogenetic analysis (29). Recombination within loci and the homoplasy it induces would be expected to increase with locus length because longer loci have more opportunities for recombination over the history of lineages. We tested for the effect of recombination by plotting the consistency index of loci, a measure of homoplasy and hence recombination, versus the length of each locus. We did not find the positive correlation expected if recombination were an important force (SI Appendix, Fig. S10). Thus, despite the higher-than-usual length of individual loci in our data, recombination appears not to be a systematically confounding factor in this data set. Additionally, long loci in this data set are advantageous for species tree estimation, a situation that does not apply to concatenation methods, where it is primarily the total number of base pairs across loci that is relevant. The average bootstrap value of each of the 447 gene trees is positively correlated with the locus length (SI Appendix, Fig. S11). Whereas the average bootstrap value of a eutherian species tree made from the longest 50 loci is 90.71, the average value for the species tree made from the 50 shortest genes is only 81.08. A similar pattern was found for analyses with the longest and shortest 100 and 200 genes sets (SI Appendix, Table S5). Thus, long loci may have contributed to the resolution of the eutherian tree using coalescent methods and may represent an efficient strategy for future phylogenomic studies.
Insufficiency of the Data of Meredith et al. (2011) for Resolving the Phylogeny of Eutherian Mammals.
One recent effort to resolve eutherian mammal phylogeny used both concatenation and coalescent methods (13), based on portions of 26 genes and extensive taxon sampling representing all eutherian families. Meredith et al. (13) suggested that coalescent methods are inappropriate for reconstructing deep-level phylogenies because they were unable to resolve even uncontroversial eutherian nodes with a high level of confidence. The data set of Meredith et al. shares 31 species with our study, representing 16 of 18 eutherian orders. We examined the capacity of the genetic data (26 genes) of Meredith et al. to resolve the phylogenetic relationships of these 31 species. For these analyses, we used two coalescent methods, MP-EST and STAR, and the concatenation method MrBayes.
As expected, both coalescent analyses produced eutherian trees that received low bootstrap support values for most of the nodes, indicating that eutherian phylogeny could not be resolved with the amount of genetic data provided (SI Appendix, Figs. S12 and S13). In the concatenation tree, by contrast, the posterior probability supports for most of nodes were equal to 1.0, even though this tree differed topologically from both the one generated by the full data set of Meredith et al. and the one generated in our study (Fig. 1). The topological incongruence indicates that the high nodal support values arising from use of the concatenation method in this case are likely spurious (SI Appendix, Fig. S14) and is consistent with our finding that taxon sampling is a confounding factor that can mislead phylogenetic results using concatenation methods. The above results indicate that the number of loci used in Meredith et al. (13) is insufficient to resolve even the reduced mammal tree with 31 taxa.
We conducted a simulation analysis to estimate the number of genes required to resolve the eutherian tree of Meredith et al. (13) with high confidence, given their extensive taxon sampling. We first estimated a MP-EST tree for the original data set of Meredith et al., including all 169 taxa and their 26 genes. This MP-EST tree has branch lengths in coalescent units, allowing us to simulate gene trees from it. Next, we simulated 25 gene trees from the MP-EST tree based on the coalescent model (30), and then the simulated gene trees were used as data to construct a MP-EST tree. The simulation was repeated 100 times, and then a consensus tree was built from the 100 MP-EST trees. We repeated the above steps by increasing the number of simulated gene trees (sample size) to 50, 75, 100, 200, 400, and 600, respectively. On the basis of the simulations, we estimate that, given their taxon sampling, Meredith et al. would require a minimum of 400 genes to achieve a species tree dominated by high-confidence nodes and a minimum bootstrap confidence of 50% (SI Appendix, Fig. S15). This estimate is a lower bound, because our simulation did not include gene tree error when estimated from DNA sequences. In addition, we calculated the average bootstrap value of the eutherian tree with 169 taxa and the original gene sampling (26 genes) of Meredith et al. (SI Appendix, Fig. S16) and compared it to that from the subsampled tree with 31 taxa (SI Appendix, Fig. S12) using the coalescent method MP-EST. We found that the average bootstrap values increase only 0.5% from 71.7 to 72.2%, indicating that the extensive species sampling did not compensate for the effect of limited gene sampling in this case. Consequently, we suggest that some phylogenetic conclusions of the concatenation analyses of Meredith et al. should be treated with caution.
Discussion
Efforts to elucidate phylogenetic relationships among eutherian mammals have been pursued intensively by increasing the sampling of taxa and/or genetic data (12–18). Controversies about some key elements of eutherian relationships, however, appear to be stubbornly irreconcilable (7–13). This study demonstrates that these controversies can at least partially be explained by the incongruence introduced by concatenation methods, which can result in misleading phylogenies. In addition, the high level of gene tree heterogeneity in this study is surprising, especially given the recent suggestion that coding sequences may be less subject to ILS than noncoding sequences due to frequent selective sweeps, which tend to remove ILS (25).
Although the mammal data set of this study is rich in the number of loci, it is not comprehensive in taxon sampling. Studies have shown that taxon sampling is an important component for accurately estimating phylogenies (31, 32). For example, a recent study in yeasts shows that increasing taxon sampling can resolve phylogenetic relationships that appear to be controversial using fewer taxa (33). The results of the present study suggest that, although taxon sampling remains important for phylogenetic analysis, it is also critical to gather sufficient numbers of loci to obtain a reliable phylogeny for eutherian mammals and other clades in the Tree of Life.
“Species tree” methods were early on recognized for yielding lower bootstrap or posterior probabilities than the corresponding analyses of the same data sets by methods using concatenation (4, 9). These results could suggest that the confidence of species trees was inaccurate, or that the confidence of concatenation studies was inflated, or both. Consistent with early empirical studies, simulations, and theory, our results suggest that overconfidence of concatenation results, whether Bayesian or likelihood, is likely operating in the mammal data set. This explanation seems the most parsimonious for explaining the pattern of incongruence among high-confidence nodes observed in our subsampling analyses of data sets. Greater attention to accurate alignments, substitution models, and nonstationarity may reduce the erratic behavior of concatenation methods (34) and improving the accuracy of individual gene trees may improve species tree estimation as well.
By accommodating gene tree heterogeneity and variable taxon sampling, the coalescent analyses reported here provide a consistent and well-resolved phylogeny for eutherian mammals (Fig. 1). Our results strongly support the Atlantogenata hypothesis of the eutherian root, suggesting that the first major eutherian diversification was caused by the separation of the Laurasia from the Gondwana (14, 16). A recent analysis using STAR based on flanking regions of ultraconserved elements recovered a tree that places Afrotheria as the most basal clade of Eutheria (35), but it is unclear how the signal in their gene trees differ from those in our analysis. In addition, our study confirms Scandentia as the sister group of Primates, providing a context to study early character and genome evolution in the lineages leading to primates and humans (36). Finally, our data support Perissodactyla (odd-toed ungulates), Carnivora, and Cetartiodactyla (including even-toed ungulates and Cetacea) as a monophyletic clade within Laurasiatheria (37–40). Differing from the traditional view, however, we find that odd-toed ungulates are more closely related to carnivores than to even-toed ungulates (38–40), suggesting an emergence of carnivores from within a paraphyletic ungulate clade. We expect the refinement and completion of eutherian phylogeny in the future as more taxa with genome-scale data become available.
The increasing availability of genome-scale data should lead to further refinements of the Tree of Life. However, the use of genomic data for increasing numbers of species constitutes a major challenge in the field of phylogenetics due to the prevalence of gene tree heterogeneity. Our study suggests that coalescent methods can provide an accurate and consistent reconstruction of species phylogenies, despite the complexities commonly observed in phylogenomic data.
Materials and Methods
Model Selection.
The best-fit substitution model for each of the 447 genes was selected by the Akaike Information Criterion (AIC). The log-likelihoods of the substitution models for 447 genes were obtained in RAxML (23) and then used to calculate AIC = −2log-likelihood + 2_P_, where P is the number of parameters in the model. It was suggested by the Akaike Information Criterion that GTR+Γ is the best-fit model for 364 genes, whereas TIM+ Γ is the best-fit model for the remaining 83 genes. Additionally, the second best-fit model for the 83 genes is GTR+Γ, and the difference of the AIC between the two models, GTR+ Γ and TIM+Γ, is less than 3. We constructed gene trees for each of those 83 genes using both GTR+ Γ and TIM+Γ models, and the gene trees based on both models are identical in topologies. Thus, GTR+ Γ was selected as the substitution model used in the concatenation and coalescent methods for reconstructing the phylogeny of eutherian mammals.
Phylogenetic Analyses.
We used two coalescent methods: MP-EST (8) and STAR (9). Details of these methods are explained in Results.
For MP-EST analysis, individual gene trees for each of the 447 loci were estimated using the maximum-likelihood method RAxML (23) and rooted by an outgroup (Chicken). Species trees were estimated from the rooted gene trees in the program MP-EST with 100 bootstrap replicates (8).
The STAR analyses were conducted using Phybase (30) with the neighbor-joining algorithm on a matrix of ranks of taxon pairs in the gene trees estimated by RAxML (23) under a GTR+Γ model and using chicken as an outgroup. The data sets for STAR analyses were bootstrapped for 100 replicates in Phybase (30). Specifically, we first resampled genes with replacement and then resampled sites with replacement for each resampled gene, as recommended (41). A STAR tree was constructed from each multilocus pseudoreplicate, and a majority rule consensus STAR tree was then built from the 100 replicates (42).
We used the Bayesian reconstruction of concatenated sequence method as implemented in the program MrBayes 3.1.2 (22, 43). We used the default priors with the substitution parameters unlinked across partitions (or genes). The analyses were conducted for 10 million generations, sampled every 1,000 generations, and two simultaneously independent runs with two chains were performed. The average SD of split frequencies was <0.01. In addition, we performed the maximum-likelihood analyses for the concatenated sequence dataset using the program RAxML (23). The maximum-likelihood estimates were bootstrapped for 100 replicates based on the GTR+ Γ substitution model.
Subsampling of Loci.
We estimated species trees and concatenation trees from subsets of the 447 loci. We selected loci at random, sampling 25, 50, 100, 200, and 300 loci. For each analysis, we selected 10 gene sets of each size as replicates. For each subsampling, we estimated species trees by MP-EST, STAR, MrBayes, and RAxML as above. We conducted bootstrapping on each subsample and then averaged the bootstrap values for relevant branches to obtain the lines in Fig. 2_A_. We also examined each of the 10 replicates to determine if a given clade received >90% bootstrap support in the case of MP-EST, STAR, or RAxML or >0.9 posterior probability in the case of MrBayes. These data were used to create the heatmap in Fig. 2_B_.
Subsampling of Taxa.
We repeated the phylogenetic analyses with two new taxon sets by excluding 6 and 12 eutherian taxa from the original taxon set. The taxa excluded were selected from each of the four eutherian superorders. The taxa excluded are provided in SI Appendix, Table S3.
Test of the Multispecies Coalescent Model.
We evaluated how well the multispecies coalescent model can explain the gene tree variation observed by simulating gene trees on the species tree estimated by MP-EST. This species tree contains branch lengths in coalescent units, which are sufficient for simulating under a standard multispecies coalescent model in the R package Phybase (30). We calculated the Robinson–Foulds distances (44) between gene trees observed in the empirical data set, as well as an expected set of distances based on the simulated data (SI Appendix). We also calculated the frequency of gene trees from triplets of taxa as a test of the multispecies coalescent model using the method outlined in Ané (27) (SI Appendix). We summarized a majority-rule consensus tree using PHYLIP v.3.69 (42, 45) for expected and observed gene trees, respectively (SI Appendix).
Supplementary Material
Supporting Information
Acknowledgments
We thank J. Meng, Z. Luo, C. Davis, F. Rheindt, and J. McCormack for comments, the Tsinghua National Laboratory for Information Science and Technology, Harvard University Research Computing Cluster, and Georgia Advanced Computing Resource for computing support. This work was supported by the startup funds of L.L. provided by the University of Georgia, by National Science Foundation Grant DEB-0743616 (to S.V.E.), by funds from the National Natural Science Foundation of China (31171047 to S.S.), and by the startup funds of S.W. provided by Shenyang Normal University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.de Queiroz A, Gatesy J. The supermatrix approach to systematics. Trends Ecol Evol. 2007;22(1):34–41. doi: 10.1016/j.tree.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 2.William J, Ballard O. Combining data in phylogenetic analysis. Trends Ecol Evol. 1996;11:334. doi: 10.1016/0169-5347(96)81133-5. [DOI] [PubMed] [Google Scholar]
- 3.Belfiore NM, Liu L, Moritz C. Multilocus phylogenetics of a rapid radiation in the genus Thomomys (Rodentia: Geomyidae) Syst Biol. 2008;57(2):294–310. doi: 10.1080/10635150802044011. [DOI] [PubMed] [Google Scholar]
- 4.Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63(1):1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- 5.Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol. 2007;56(1):17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- 6.Mossel E, Vigoda E. Phylogenetic MCMC algorithms are misleading on mixtures of trees. Science. 2005;309:2207–2209. doi: 10.1126/science.1115493. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Pearl DK, Brumfield RT, Edwards SV. Estimating species trees using multiple-allele DNA sequence data. Evolution. 2008;62:2080–2091. doi: 10.1111/j.1558-5646.2008.00414.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Yu L, Edwards SV. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol Biol. 2010;10:302. doi: 10.1186/1471-2148-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Yu L, Pearl DK, Edwards SV. Estimating species phylogenies using coalescence times among sequences. Syst Biol. 2009;58:468–477. doi: 10.1093/sysbio/syp031. [DOI] [PubMed] [Google Scholar]
- 10.Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Murphy WJ, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 12.Murphy WJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 13.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 14.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolaev S, et al. NISC Comparative Sequencing Program Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 2007;3:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wildman DE, et al. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci USA. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad AB, Allard MW, Green ED, Green ED, NISC Comparative Sequencing Program Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kriegs JO, et al. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallström BM, Kullberg M, Nilsson MA, Janke A. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol Biol Evol. 2007;24:2059–2068. doi: 10.1093/molbev/msm136. [DOI] [PubMed] [Google Scholar]
- 20.Asher RJ, Bennett N, Lehmann T. The new framework for understanding placental mammal evolution. Bioessays. 2009;31:853–864. doi: 10.1002/bies.200900053. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Yu L, Kubatko L, Pearl DK, Edwards SV. Coalescent methods for estimating phylogenetic trees. Mol Phylogenet Evol. 2009;53:320–328. doi: 10.1016/j.ympev.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 22.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 23.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki Y, Glazko GV, Nei M. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc Natl Acad Sci USA. 2002;99:16138–16143. doi: 10.1073/pnas.212646199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobolth A, Dutheil JY, Hawks J, Schierup MH, Mailund T. Incomplete lineage sorting patterns among human, chimpanzee, and orangutan suggest recent orangutan speciation and widespread selection. Genome Res. 2011;21:349–356. doi: 10.1101/gr.114751.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ané C. Reconstructing concordance trees and testing the coalescent model from genome-wide data sets. In: Knowles LL, Kubatko L, editors. Estimating Species Trees: Practical and Theoretical Aspects. Wiley-Blackwell, Hoboken, NJ: 2010. pp. 35–52. [Google Scholar]
- 28.Nishihara H, Okada N, Hasegawa M. Rooting the eutherian tree: The power and pitfalls of phylogenomics. Genome Biol. 2007;8:R199. doi: 10.1186/gb-2007-8-9-r199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castillo-Ramirez S, Liu L, Pearl DK, Edward SV. Bayesian estimation of species trees: A practical guide to optimal sampling and analysis. In: Knowles LL, Kubatko L, editors. Estimating Species Trees: Practical and Theoretical Aspects. Wiley-Blackwell, Hoboken, NJ: 2010. pp. 15–33. [Google Scholar]
- 30.Liu L, Yu L. Phybase: An R package for species tree analysis. Bioinformatics. 2010;26:962–963. doi: 10.1093/bioinformatics/btq062. [DOI] [PubMed] [Google Scholar]
- 31.Hillis DM, Pollock DD, McGuire JA, Zwickl DJ. Is sparse taxon sampling a problem for phylogenetic inference? Syst Biol. 2003;52:124–126. doi: 10.1080/10635150390132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollock DD, Zwickl DJ, McGuire JA, Hillis DM. Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol. 2002;51:664–671. doi: 10.1080/10635150290102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 34.Philippe H, et al. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCormack JE, et al. Untraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species tree analysis. Genome Res. 2011 doi: 10.1101/gr.125864.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janecka JE, et al. Molecular and genomic data identify the closest living relative of primates. Science. 2007;318:792–794. doi: 10.1126/science.1147555. [DOI] [PubMed] [Google Scholar]
- 37.Roca AL, et al. Mesozoic origin for West Indian insectivores. Nature. 2004;429:649–651. doi: 10.1038/nature02597. [DOI] [PubMed] [Google Scholar]
- 38.Arnason U, et al. Mitogenomic relationships of placental mammals and molecular estimates of their divergences. Gene. 2008;421(1–2):37–51. doi: 10.1016/j.gene.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 39.Amrine-Madsen H, Koepfli KP, Wayne RK, Springer MS. A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol Phylogenet Evol. 2003;28:225–240. doi: 10.1016/s1055-7903(03)00118-0. [DOI] [PubMed] [Google Scholar]
- 40.Waddell PJ, Okada N, Hasegawa M. Towards resolving the interordinal relationships of placental mammals. Syst Biol. 1999;48(1):1–5. [PubMed] [Google Scholar]
- 41.Seo TK. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Mol Biol Evol. 2008;25:960–971. doi: 10.1093/molbev/msn043. [DOI] [PubMed] [Google Scholar]
- 42.Margush T, McMorris FR. Consensus n-trees. Bull Math Biol. 1981;43:239–244. [Google Scholar]
- 43.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 44.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math Biosci. 1981;53:131–147. [Google Scholar]
- 45.Felsenstein J. PHYLIP: Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 46.Swofford D. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). version 4. Sinauer Associate (Sunderland, MA)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information