Nuclear pore component Nup98 is a potential tumor suppressor and regulates post-transcriptional expression of select p53 target genes (original) (raw)
. Author manuscript; available in PMC: 2013 Dec 14.
Summary
The p53 tumor suppressor utilizes multiple mechanisms to selectively regulate its myriad target genes, which in turn mediate diverse cellular processes. Here, using conventional and single molecule mRNA analyses, we demonstrate that the nucleoporin Nup98 is required for full expression of p21, a key effector of the p53 pathway, but not several other p53 target genes. Nup98 regulates p21 mRNA levels by a post-transcriptional mechanism in which a complex containing Nup98 and the p21 mRNA 3′-UTR protects p21 mRNA from degradation by the exosome. An in silico approach revealed another p53 target (14-3-3σ) to be similarly regulated by Nup98. The expression of Nup98 is reduced in murine and human hepatocellular carcinomas (HCC) and correlates with p21 expression in HCC patients. Our study elucidates a previously unrecognized function of wild-type Nup98 in regulating select p53 target genes that is distinct from the well-characterized oncogenic properties of Nup98 fusion proteins.
Introduction
The p53 tumor suppressor functions as a sequence-specific transcriptional regulator for a great variety of target genes. Different subsets of these target genes mediate various cellular outcomes such as cell cycle arrest, senescence, programmed cell death or others (reviewed in (Vousden and Prives, 2009). The underlying molecular processes that dictate p53 target gene preference are complex and still only partially understood. Previously we found that the nuclear transport factor hCAS/Cse1L, binds to a subset of p53 target gene promoters and is required for their full activation upon stress-mediated p53 induction (Tanaka et al., 2007).
An RNAi screen against other nuclear transport factors, nuclear pore components in particular, suggested that the nucleoporin Nup98 may also contribute to target gene selectivity of p53. Nucleoporins (Nups), essential components of the nuclear transport machinery, form the nuclear pore complex (NPC), which is embedded in the nuclear envelope and facilitates nucleocytoplasmic trafficking (D'Angelo and Hetzer, 2008). An increasing body of evidence (largely from studies in yeast) suggests that in addition to their well-characterized role in transport, some Nups are also involved in regulating gene expression (Kohler and Hurt, 2010; Strambio-De-Castillia et al., 2010). In particular, Nup98, a major component of the NPC (Cronshaw et al., 2002) was reported to move from the NPC into the nucleoplasm in a transcription-dependent manner (Griffis et al., 2002) and also to be involved in mRNA export (Enninga et al., 2002; Powers et al., 1997), Further, Drosophila Nup98 is required for activation of genes related to stress response, cell cycle progression, and development (Capelson et al., 2010; Kalverda et al., 2010). Nup98 has also been linked to cancer, yet only within the context of Nup98 oncogenic fusion proteins that occur as a consequence of chromosomal translocations in leukemias (e.g. Nup98-HOXA9 t(7;11)(p15;p15); Moore et al., 2007; Xu and Powers, 2009)). The function of wild-type Nup98 in tumor development is largely unknown.
Results
Nup98 Ablation Selectively Reduces p21 mRNA and Protein Accumulation Upon p53 Activation
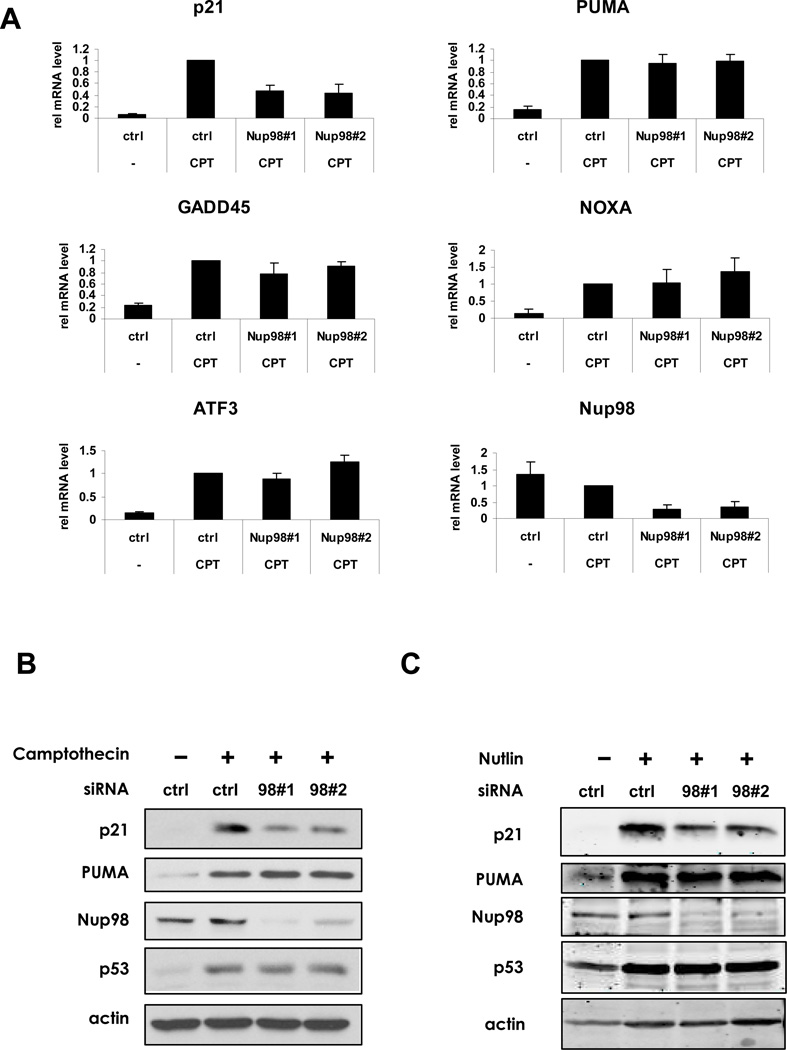
Since a Nup siRNA screen identified Nup98 as a likely determinant of p53 target gene selectivity (Figure S1A), we transfected HepG2 liver cancer cells (wild-type p53) with Nup98 siRNAs and then treated the cells with Camptothecin (CPT) to activate p53. The induction of p53 target gene mRNAs including p21, PUMA, GADD45, NOXA and ATF3 was measured by quantitative real-time PCR (qRT-PCR) (Figure 1A). p21 mRNA (Figure 1A) or protein (Figure 1B) was unique in being affected by Nup98 ablation. Similar results were obtained in HepG2 cells treated with Daunorubicin (data not shown) as well as in Huh-6 and Sk-Hep1 cells treated with CPT (Figure S1B). Nup98 knockdown also led to decreased p21 (but not PUMA) protein upon treatment with Nutlin, a compound that disrupts the p53-Mdm2 complex. This indicates that the effects of Nup98 depletion were independent of DNA damage signaling (Figure 1C). Further, Nup98 depletion selectively reduced p21 mRNA induction in Hep3B-4Bv cells that harbor a temperature-sensitive p53 (Friedman et al., 1997) at 32°C when p53 is in wild-type conformation (Figure S1C). The basal levels of p21 mRNA in the presence or absence of Nup98 were largely unchanged in three different liver cancer cell lines, suggesting that only induced p21 expression (here in response to up-regulated levels of p53) relies on full expression of Nup98 (Figure S1D). Taken together, our data suggest that Nup98 is required for induction of p21 by p53 in a DNA damage-independent manner.
Figure 1. Nup98 Depletion Selectively Reduces p21 mRNA and Protein Accumulation upon p53 Activation.
(A) HepG2 cells were treated either with control (ctrl) or two Nup98 siRNAs (Nup9898#1 and Nup9898#2) for 72 h and CPT (300 nM) was added for the final 24 h before lysis of cells for mRNA and protein analysis. Relative expression of p53 target genes: p21, PUMA, GADD45, NOXA, ATF3 and Nup98 was measured by qRT-PCR and normalized to 1 for each ctrl siRNA and drug treatment condition. Data are presented as mean ± standard deviation (SD) of three independent experiments.
(B) HepG2 cell were treated as in (A) and cell extracts were examined by immunoblotting with indicated antibodies.
(C) HepG2 cells were treated either with control (ctrl) or Nup9898#1 and Nup9898#2 siRNA for 72 h and Nutlin-3 (20 µM) was added 24 h prior to extraction of cells for protein analysis. Cell extracts were analyzed by immunoblotting with the indicated antibodies.
The predominant biogenesis of Nup98 occurs by autocleavage of a Nup98/Nup96 precursor protein (Fontoura et al., 1999; Rosenblum and Blobel, 1999). Arguing strongly against the possibility that Nup96 is also involved in p21 expression, is that, upon transfection of two Nup98 siRNAs, despite decreased amounts of precursor Nup98/96 mRNA, Nup96 protein levels remained unaltered. (Figure S1E: left panels, top and bottom; see right panel for Nup96 antibody specificity). Having excluded Nup96 as a factor in controlling p21 gene expression, we went on to examine the mode by which Nup98 regulates p21 mRNA accumulation.
Nup98 Regulates p21 by a Post-Transcriptional Mechanism
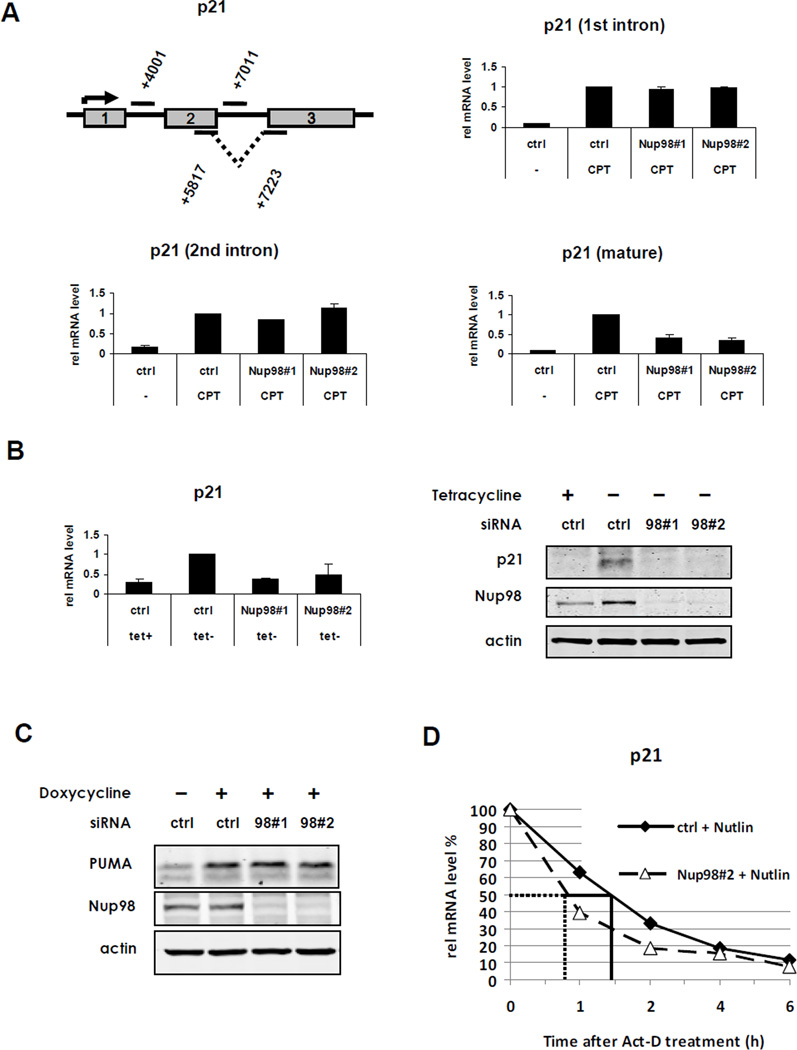
To determine the stage of p21 gene expression that is regulated by Nup98 we compared induction of nascent unspliced and mature spliced p21 mRNA in HepG2 cells, using primers that anneal to intronic regions of unspliced p21 mRNA (1st and 2nd intron, respectively) or exon-spanning primers that detect only spliced, mature p21 mRNA (Figure 2A, upper left panel). Interestingly, while induction of nascent p21 mRNA (upon CPT treatment) was not significantly impacted by Nup98 knockdown (Figure 2A, upper right and lower left panels), levels of mature p21 mRNA were significantly decreased upon Nup98 depletion compared to the control siRNA treated cells (Figure 2A, lower right panel).
Figure 2. Post-transcriptional Regulation of p21 by Nup98.
(A) Nup98 regulates the levels of mature but not nascent p21 mRNA. Upper left panel: schematic depicts primers used to amplify different regions of nascent and mature p21 mRNA. The numbers indicate the position of the amplicon within the p21 gene locus. Rectangles represent exons 1 to 3. HepG2 cells were treated as described in Figure 1A. Nascent p21 mRNA induction was measured by qRT-PCR with primer pairs annealing to intronic regions of the unspliced transcript: first intron (+4001) (upper right panel) and second intron (+7011) (lower left panel), respectively. Lower right panel: spliced p21 mRNA was measured by using the exon-spanning primer pair (+5817/+7223) indicated in the schematic. Data are presented as mean ± SD of two independent (experiments.
(B) Nup98 regulates exogenous p21 mRNA. Left panel: H24-p21 cells containing a tetracycline (Tet-off)-regulatable p21 expression construct were treated either with control (ctrl) or Nup98 siRNAs (Nup9898#1 and Nup9898#2) for 72 h. as in Figure 1. p21 was induced by removal of Tet 24 h before harvesting. Left panel shows relative p21 and Nup98 mRNA expression measured by qRT-PCR and normalized to 1 for the “ctrl siRNA tet-“ condition. Data are presented as mean ± SD derived from two independent experiments. Right panel: protein expression from the same experimental condition analyzed by immunoblotting with indicated antibodies.
(C) PUMA levels are not affected by Nup98 knock-down H1299-derivative cell line containing a tetracycline (Tet-on)-regulatable PUMA expression construct (H24-PUMA) was treated either with control (ctrl) or two Nup98 siRNAs (Nup9898#1 and Nup9898#2) for 72 h. PUMA was induced by adding Doxycycline 24 h before harvesting. Cell extracts were analyzed by immunoblotting with indicated antibodies.
(D) Nup98 stabilizes mature p21 mRNA HepG2 cells were treated either with control (ctrl) or Nup9898#2 siRNA (Nup9898#2) for 72 h and Nutlin-3 (20 µM) was added 24 h before blocking mRNA synthesis with 0.4 µM Actinomycin-D (Act-D). Cells were harvested at indicated time points and mature p21 mRNA decay was measured by qRT-PCR with the exon-spanning primer pairs described in Figure 2A. The solid and dotted lines indicate the half-life of p21 in ctrl-siRNA or Nup9898#2 conditions, respectively.
To confirm that the lowered p21 mRNA levels upon Nup98 knockdown were independent of transcription initiation, and also to exclude a splicing defect, we utilized an H1299 derivative p53-null cell line (H24-p21) in which p21 mRNA is transcribed from a p21 cDNA construct under the control of an artificial “tet-off” promoter (Niculescu et al., 1998). This construct lacks intronic regions, but does contain part of the p21 3’-UTR. In these cells Nup98 knockdown again resulted in impaired accumulation of p21 mRNA (Figure 2B left panel) and protein (Figure 2B right panel). As before, Nup96 protein levels were unaltered by Nup98 siRNAs (Nup98#1 and Nup9898#2) despite a decrease in the Nup98/Nup96 precursor mRNA (Figure S2A). In a related H1299 cell derivative, inducible ectopic PUMA expression was unaltered by Nup98 knockdown (Figure 2C).
Based on these findings we hypothesized that Nup98 stabilizes mature p21 mRNA. To test this, we performed mRNA half-life experiments by blocking transcription with Actinomycin-D in the presence or absence of Nup98 siRNAs. As predicted, p21 mRNA induced either by Nutlin (Figure 2D) or CPT (Figure S2B) treatment in HepG2 cells had a shorter half-life (approximately 45 minutes) when Nup98 was ablated by siRNA (siNup9898#2) compared to the control condition (approximately 90 minutes). Similar findings were obtained with another Nup98 siRNA (Figure S2C). Consistent with the observation that uninduced p21 mRNA levels do not change in response to Nup98 ablation in HepG2 cells, we did not observe differences in basal p21 mRNA half-life under these conditions (Figure S2D).
Taken together, Nup98-mediated regulation of p21 mRNA occurs by a splicing-independent post-transcriptional mechanism that involves stabilization of the mature p21 mRNA transcript.
Decreased p21 mRNA Accumulation upon Nup98 Knockdown Occurs in the Nucleus and the Cytoplasm and can be Rescued by Blocking the Exosome
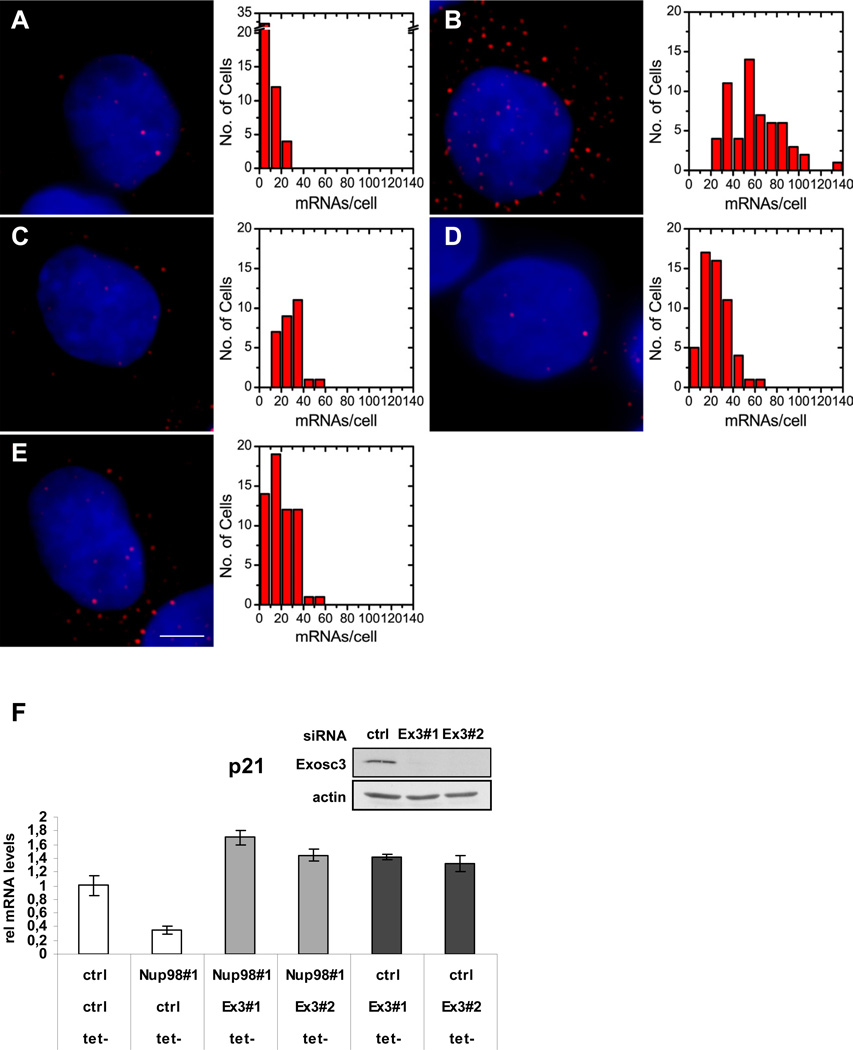
Our results indicated that Nup98 knockdown is associated with a higher rate of p21 mRNA turnover. Since mRNA degradation can occur at each step from the site of transcription to the site of translation (Houseley and Tollervey, 2009), we sought to determine in which cellular compartment Nup98 affected p21 mRNA stability. Furthermore, since Nup98 is reported to be involved in mRNA export (Enninga et al., 2002) we wanted to address whether or not Nup98 knockdown leads to nuclear p21 mRNA accumulation. We used Single Molecule Sensitive Fluorescence in situ Hybridization (mRNA FISH) staining (Femino et al., 1998), to obtain a detailed and quantitative view of nuclear and cytoplasmic p21 mRNA molecules before and after p53 activation in the presence or absence of Nup98. As expected, Nutlin treatment caused an increase in the overall number of single p21 mRNA transcripts in HepG2 cells (Figure 3A-B and summarized in Table S1) which was significantly decreased upon Nup98 knockdown (Figure 3C-D and Table S1). Nup98 depletion led to a marked reduction in cytoplasmic and more modest diminution of nuclear p21 mRNA molecules in Nutlin treated cells (Figure 3C-D and Table S1). As an additional control, in cells containing p21 siRNA even fewer cytoplasmic p21 mRNA molecules were detected (Figure 3E and Table S1). Although p21 mRNA did not accumulate in the nucleus upon Nup98 depletion (but actually decreased), a role for Nup98 in p21 mRNA export remains plausible (see Discussion section).
Figure 3. Nup98 Affects Nuclear and Cytoplasmic p21 mRNA level and Protects p21 mRNA from Degradation by the Exosome.
Nup98 knock-down affects the levels of nuclear and cytoplasmic p21 mRNA.
(A-E). Left panels show images of HepG2 cells that were treated either with ctrl (A,B) or Nup9898#1 (C) Nup9898#3 (D) or p21 (E) siRNAs for 72 h and then treated with either DMSO (A) or Nutlin-3 (20 µM; B,C,D and E) for 24 h prior to single-mRNA FISH analysis as described in Methods. FISH images were detected in the Cy5 channels and color-coded as red. The blue channel contains a DAPI stain to demarcate the nuclei. For display 5 to 15 images, depending on cell shape, from the cell mid-section were averaged in both channels. Scale bar shown in panel E equals 2 µm in all images. Right panels are histograms of p21 mRNA molecule counts per cell corresponding to panels A (48 cells counted); B (58 cells counted), C (29 cells counted) D (55 cells counted) and E (59 cells counted). Data are pooled from three repeats of the experiments with errors and statistical analysis shown in Table S1.
(F) Exosc3 knockdown rescues lowered p21 mRNA levels upon Nup98 depletion. H24 p21 tet-off cells were treated individually with either control (ctrl) or Nup9898#1 siRNA, or co-transfected with Exosc3 siRNA (Ex398#1 and Ex398#2) for 72 h as indicated. p21 was induced by removal of Tet 48 h before harvesting. Relative p21 mRNA levels were measured with qRT-PCR. Data are presented as mean ± SD (left panel). Western blot (inset) shows Exosc3 levels in p21 H24 cells treated either with ctrl or Ex398#1 and Ex398#2 siRNA. Actin serves as loading control.
mRNA degradation in yeast strains with mutated Nup116 (a homologue of mammalian Nup98) is mediated by the exosome (Das et al., 2003). We therefore determined whether the reduced levels of p21 mRNA could be rescued by co-depletion of Exosc3 (Rrp40), a core component of the exosome. As shown in Figure 3F, co-depletion of Exosc3 with Nup98 in the H24-p21 cell line prevented the decrease of p21 mRNA levels observed after Nup98 knockdown alone. These results implicate a role of Nup98 in stabilizing p21 mRNA transcripts in a manner that precludes their turnover by the exosome.
The 3′-UTR of p21 mRNA Associates with Nup98 and is Required for Nup98-Mediated Increased p21 mRNA Levels
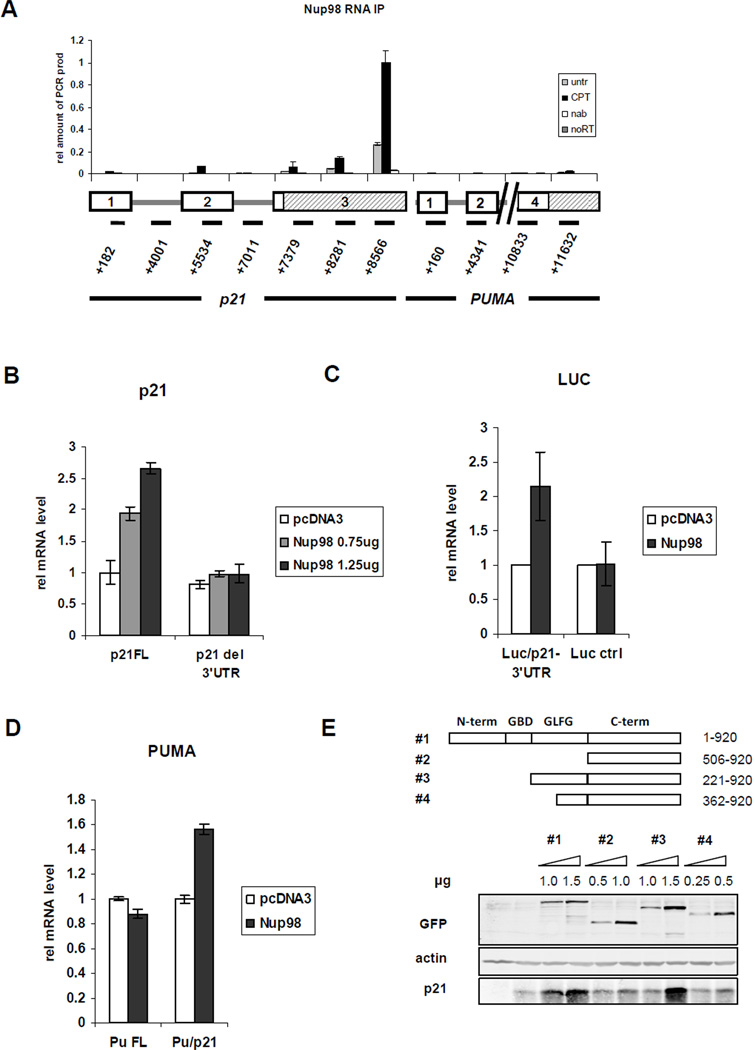
Nup98 localizes primarily at the NPC, reflected in a punctate nuclear rim staining, and to a minor extent within the nucleus, detected as intranuclear foci previously described as GLFG bodies (Enninga et al., 2002; Griffis et al., 2002; and Figure S3A). We hypothesized that Nup98 may associate with p21 mRNA and thereby affect its stability in the nucleus (and the cytoplasm as well). To address this, RNA-IP experiments were performed in which Nup98 was immunoprecipitated from formaldehyde cross-linked samples of HepG2 cells that were untreated or CPT-treated (Figure 4A) or treated with Nutlin (data not shown). For amplification by qRT-PCR, primer pairs specific for different regions of p21 mRNA or PUMA mRNA (as a negative control) were used to localize the potential regions of interaction with Nup98 (or Nup98-associated protein). Remarkably, Nup98 could be co-immunoprecipitated with p21 mRNA with the most significant interaction being detected in the 3′-UTR. By contrast, an anti-Nup98 RNA-IP revealed almost no interaction within the PUMA 3’-UTR. Nup98 also co-immunoprecipitated with p21 mRNA in H24 p21 tet-off cells, again with the strongest interaction being detected in the 3’-UTR (Figure S3B).
Figure 4. The 3’-UTR of p21 mRNA Co-associates with Nup98 and Is Required for Nup98 Mediated Increased Levels of p21 mRNA.
(A) Nup98 associates with the 3’-UTR of p21. RNA-IP was performed as described in Methods. Briefly, Nup98 was immunoprecipitated from 0.1% formaldehyde cross-linked samples of CPT (300 nM for 24 h) treated or untreated HepG2 cells. Co-immunoprecipitated mRNAs cross-linked to Nup98 were reverse-transcribed and amplified by qRT-PCR. Primer pairs specific for different regions of p21 mRNA or PUMA mRNA were used. In the schematic below the graph rectangles represent exonic regions of the respective genes and shaded rectangles indicate the 3’-UTRs of p21 and PUMA mRNA as indicated. The numbers below the lines indicate the position of each amplicon within the p21 or PUMA gene locus, respectively. Data are presented as mean ± SD.
(B) The 3’-UTR of p21 mRNA is required for increased p21 expression levels mediated by ectopically expressed Nup98. Hep3B cells were co-transfected with HA-tagged Nup98 (0.75 or 1.25 µg) and p21 cDNA containing the 3’-UTR (p21 FL, 200 ng) or p21 cDNA without the 3’-UTR (p21 del 3’-UTR, 20ng) 24h prior to RNA extraction. p21 mRNA levels transcribed from the respective constructs were measured with qRT-PCR. Data are presented as mean ± SD.
(C) The 3’-UTR of p21mRNA is sufficient for stabilization by Nup98. H1299 cells were co-transfected with HA-tagged Nup98 (1.75 µg) and a Luciferase construct (Luc/p21 3’UTR, 200 ng) containing the 3’UTR of p21 mRNA or the Luciferase construct lacking the p21 mRNA 3’UTR (Luc ctrl, 20ng) 48 h prior to RNA extraction. The mRNA levels transcribed from the respective constructs were measured with qRT-PCR. Data are presented as mean ± SD of two independent experiments.
(D) The 3’-UTR of p21mRNA is sufficient for stabilization by Nup98. H1299 cells were co-transfected with HA-tagged Nup98 (1.75 µg) and a construct containing the coding DNA sequence (CDS) and the 3’UTR of PUMA (Pu FL, 200ng) or a hybrid construct containing the PUMA CDS fused to the 3’UTR of p21 (Pu/p21, 200ng) The mRNA levels transcribed from the respective constructs were measured with qRT-PCR. Data are presented as mean ± SD.
(E) The GLFG repeat domain of Nup98 is required for maximal increase of p21 expression H1299 cells were co-transfected with GFP tagged full length (98#1) or truncated (98#2–4) Nup98 constructs with indicated concentrations and 1 µg untagged full length p21 cDNA 24 h before protein extraction. Immunoblot shows protein expression of transfected Nup98 and p21 detected with the indicated antibodies. Actin serves as loading control.
These results were extended by experiments showing that ectopic Nup98 was able to significantly increase the levels of co-expressed full-length p21 cDNA (p21 FL) but not p21 cDNA lacking the 3′-UTR (p21 del 3′-UTR) (Figure 4B; see Figure S3C for corresponding protein levels). Approximately 10-fold more of the transfected p21 FL construct than of the p21 del 3′-UTR construct was required to reach equivalent p21 mRNA levels, suggesting that the p21 3′-UTR per se is a negative regulator of p21 mRNA stability. We also observed that the p21 3’UTR fused to the coding DNA sequence (CDS) of Luciferase (Luc/p21 3’UTR) mediates a similar response to Nup98 (Figure 4C). Finally, Nup98 overexpression led to an increase of a hybrid construct (CDS of PUMA attached to the 3’UTR of p21; Pu/p21) while full length Puma 3’UTR (Pu FL) was unaffected (Figure 4D). Taken together our data show that the p21 3’ UTR is necessary and sufficient for the ability of Nup98 to augment the accumulation of p21 mRNA.
Two domains of Nup98 have been implicated for its role in mRNA export. One is the glycine-leucine-phenylalanine-glycine (GLFG) repeat domain that is important for transcription-dependent mobility of Nup98 and which serves as a docking site for the mRNA export receptor TAP/NXF1(Blevins et al., 2003; Griffis et al., 2004). The other is the Gle-2-binding domain (GBD) mediating the interaction of Nup98 with the mRNA export factor Rae1/Gle2 that also interacts with TAP (Blevins et al., 2003). Different truncated versions of Nup98 were expressed in H1299 cells along with co-transfected p21 (p21 FL). Overexpression of full length Nup98 (construct #1) maximally increased p21 protein expression in a concentration-dependent manner, whereas the C-terminus alone (construct #2; 506–920) only produced a modest increase (Figure 4E). Overexpression of the full length GLFG repeat domain plus the C-terminus (construct #3; 221–920) but lacking the Gle2-binding-domain (GBD) was sufficient to increase p21 protein to similar levels as did wild type Nup98. The C-terminal half of the GLFG repeat domain plus the C-terminus (construct #4; 362–920) was not able to fully stabilize p21. Thus the intact GLFG-repeat but not the GBD of Nup98 is necessary for full stabilization of p21. Nup98-dependent regulation of p21 may require the mobility of Nup98 and/or its interaction with TAP but is independent of its interaction with Rae1/Gle2. In line with this, in our siRNA screen the depletion of Rae1/Gle2 did not lead to reduced p21 mRNA accumulation (Figure S1A).
An In Silico Approach Identifies _14-3-3_σ as Another p53 Target Gene mRNA that is Bound and Regulated by Nup98
Since the strongest association of Nup98 with p21 mRNA occurs in its 3′-UTR, we sought to determine whether other mRNAs within the p53 transcriptional regulon might also be regulated post-transcriptionally by Nup98 (Keene, 2007). We adapted a computational method previously used to infer mechanisms of post-transcriptional messenger RNA stability regulation from genome-wide changes in steady-state transcript abundance in yeast (Foat et al., 2005). The REDUCE software suite (see Figure 5A and Experimental Procedures) was used to discover RNA sequence motifs whose occurrence in 3′-UTRs is predictive of the difference in gene expression 72 h after doxorubicin treatment, i.e. between cells in which p53 is active vs. inactive (Chau et al., 2009). This yielded a single, highly significant C-rich motif (Figure 5A), occurrences of which in the 3′-UTR are predictive of mRNA stabilization upon p53 activation (p=value = 5x10−26). To determine whether the motif indicates a potential mRNA or genomic DNA binding element, we analyzed how the correlation with differential gene expression differed when counting motif occurrences in 400 nt windows immediately upstream or downstream of poly-adenylation sites in genomic DNA (Figure 5B). The correlation was significantly higher for the upstream window, suggesting that the C-rich motif represents the specificity of an RNA-binding factor. The C-rich motif is also robust in the sense that using other differential steady-state expression data yields consistent results. First, occurrences in the 3′-UTR correlated significantly with the difference in mRNA expression 72 h vs. 48 h after doxorubicin treatment in p53-active cells (p = 3*10−49) (Chau et al., 2009). Second, they correlate with the difference in mRNA expression 24 h after Nutlin vs. mock treatment (p = 3*10−32) (Goldstein et al.).
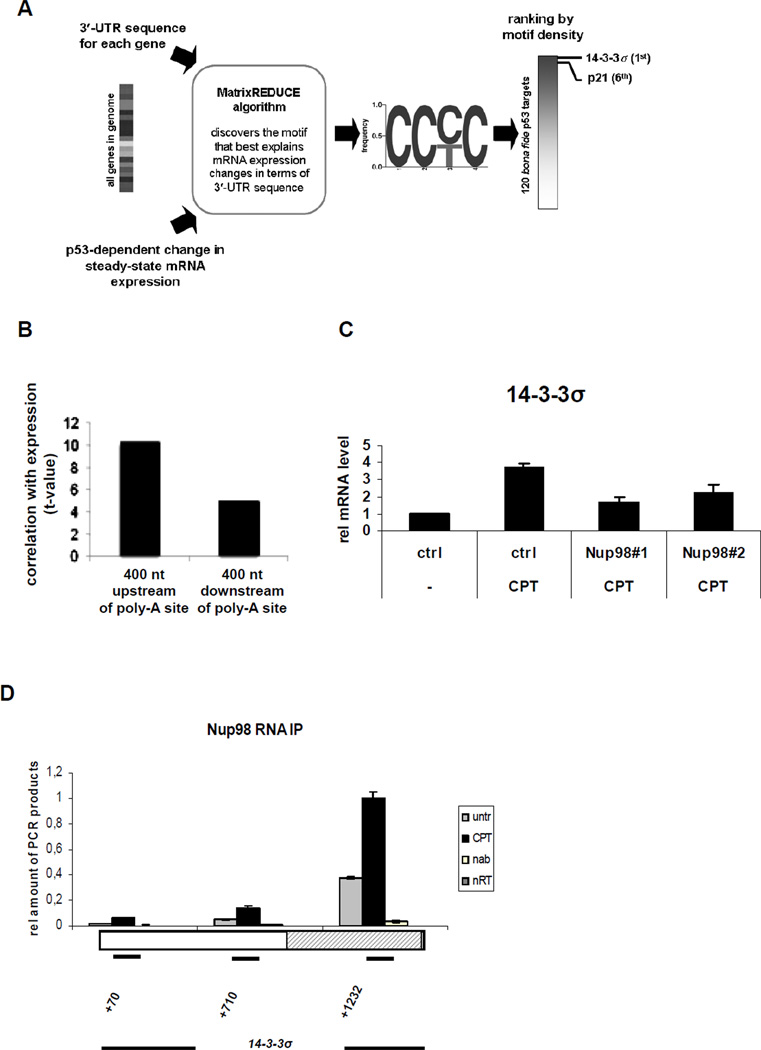
Figure 5. An In Silico Approach Identifies 14-3-3σ as another p53 Target Gene mRNA that Associates with and is Regulated by Nup98.
(A) Flow-chart describing the unbiased genomewide in silico cis-regulatory analysis that yielded the CCc/tC motif. The MatrixREDUCE algorithm was used to discover a nucleotide sequence motif (in the form of a weight matrix) whose occurrences in the 3’-UTR optimally predict p53-dependent changes in steady-state mRNA expression 72 h after doxorubicin treatment.
(B) Motif occurrences in a 400 bp window immediately upstream of the polyadenylation site are significantly more predictive of the expression changes than those downstream, suggesting that the C-rich motifs represents the specificity of an RNA-binding factor.
(C) HepG2 cells were treated either with ctrl or Nup9898#1 and Nup9898#2 siRNA for 72 h and 300 nM (CPT) was added for the final 24 h before RNA extraction. 14-3-3σ mRNA was measured by qRT-PCR. The ctrl siRNA and no CPT condition was taken as 1. Data are presented as mean ± SD of two independent experiments.
(D) RNA-IP experiment in HepG2 cells was performed as in Figure 4A. Primer pairs specific for different regions of 14-3-3σ mRNA were used to localize the potential regions of interaction with Nup98. The rectangle represents the single 14-3-3σ exon and the shaded area indicates the 3’-UTR. The numbers indicate the position of each amplicon within the 14-3-3σ gene locus. Data are presented as mean ± SD.
The above results suggested that the C-rich motif mediates a post-transcriptional and p53-dependent regulatory signal. We used this motif to rank a list of 120 bona fide p53 target genes (Riley et al., 2008) in terms of the total motif score in their 3′-UTR (Table S2). We found that 14-3-3σ scored the highest and p21 ranked 6th. Indeed, in HepG2 cells Nup98 was required for full 14-3-3σ expression (Figure 5C). Furthermore, as with p21, we were able to co-immunoprecipitate the 14-3-3σ mRNA 3′-UTR with Nup98 (Figure 5D). The finding that the highest-scoring candidate behaved similarly to p21 supports the efficacy of this in silico screen, and increases further the likelihood that Nup98 interaction with 3’UTR regions of select p53 target genes contributes to their stability.
Nup98 Depletion Increases Camptothecin-Induced Cell Death and Decreases Senescence Mediated by Nutlin
Although p21 is well-known for its ability to regulate the cell cycle, it also functions to inhibit cell death under stress conditions (reviewed in (Abbas and Dutta, 2009). Decreased p21 levels observed after Nup98 knockdown may sensitize cells to drug induced apoptosis. To test this, HepG2 cells were treated with siRNAs specific for Nup98 or p21 and harvested after 24 h treatment with CPT followed by FACS analysis. While there were no major changes in cell cycle distribution, there was an increase in cell death after Nup98 knockdown (Figure 6A). This corresponded well with the results of p21 siRNA-mediated knockdown where we observed an even higher percentage of sub-G1 cells, possibly due to an even greater reduction of p21 protein levels (Figure 6B). Decreased p21 induction associated with Nup98 depletion is very likely to be a contributing factor to the increased cell death that occurs upon Nup98 knockdown.
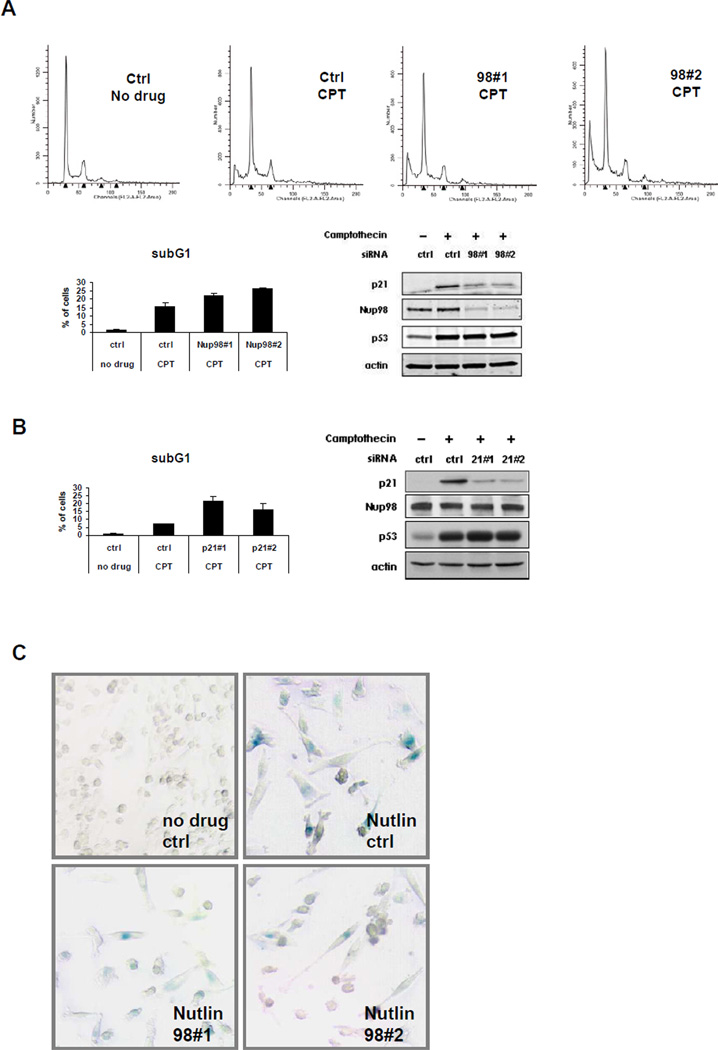
Figure 6. Nup98 Depletion Increases Camptothecin (CPT) Induced Cell Death and Decreases Nutlin Induced Senescence.
(A) Nup98 knock-down sensitizes cells to CPT induced apoptosis. Upper panels: FACS analysis was performed with propidium iodide stained samples of HepG2 cells following transfection with control (ctrl) or Nup98 (9898#1 and 9898#2) siRNAs and drug treatment as described in Figure 1A. Data was analyzed using the ModFit program. Lower left panel: bar diagram shows relative % of subG1 cells taken as a measurement of cell death for each indicated condition. Data are presented as mean ± SD derived from two independent experiments. Lower right panel: protein levels in HepG2 cells treated as above were analyzed by immunoblotting with indicated antibodies.
(B) p21 knockdown recapitulates the effects of Nup98 knockdown in CPT treated cells. Left panel: FACS analysis was performed in HepG2 cells as described in (A) following transfection with p21 siRNA (2198#1 and 2198#2) and drug treatment. Bar diagram shows relative percentage (%) of subG1 cells as a measurement of cell death for each indicated condition. Data are presented as mean ± SD derived from two independent experiments. Right panel: protein levels in cells treated as above detected by Western blotting with indicated antibodies.
(C) Nup98 knock-down decreases Nutlin induced senescence. Sk-Hep1 cells were treated either with ctrl or Nup9898#1 and Nup9898#2 siRNA for 24h and Nutlin-3 (10 µM) was added as indicated 5 days prior to staining senescence associated β galactosidase (SA- β -Gal). Shown are representative images of cells with SA-β -Gal staining for the indicated conditions.
Cellular senescence is another important p53-mediated and p21-related cellular outcome (Abbas and Dutta, 2009). After 5 days of Nutlin treatment of control siRNA transfected Sk-Hep1 cells, there was a strong increase of senescence-associated beta-galactosidase activity (SA-beta-Gal) (43±15% positive cells) compared to corresponding DMSO treated controls (<0.01% positive cells) which was significantly diminished when Nup98 levels were reduced (12±9% and 16±8%, in cells containing Nup98 siRNA #1 and siRNA98#2, respectively) (Figure 6C). Thus Nup98 offers partial protection from DNA damage mediated apoptosis, but cells require Nup98 to undergo senescence mediated by p53.
Nup98 Expression is Downregulated in an HCC Mouse Model and in Human Patients with HCC
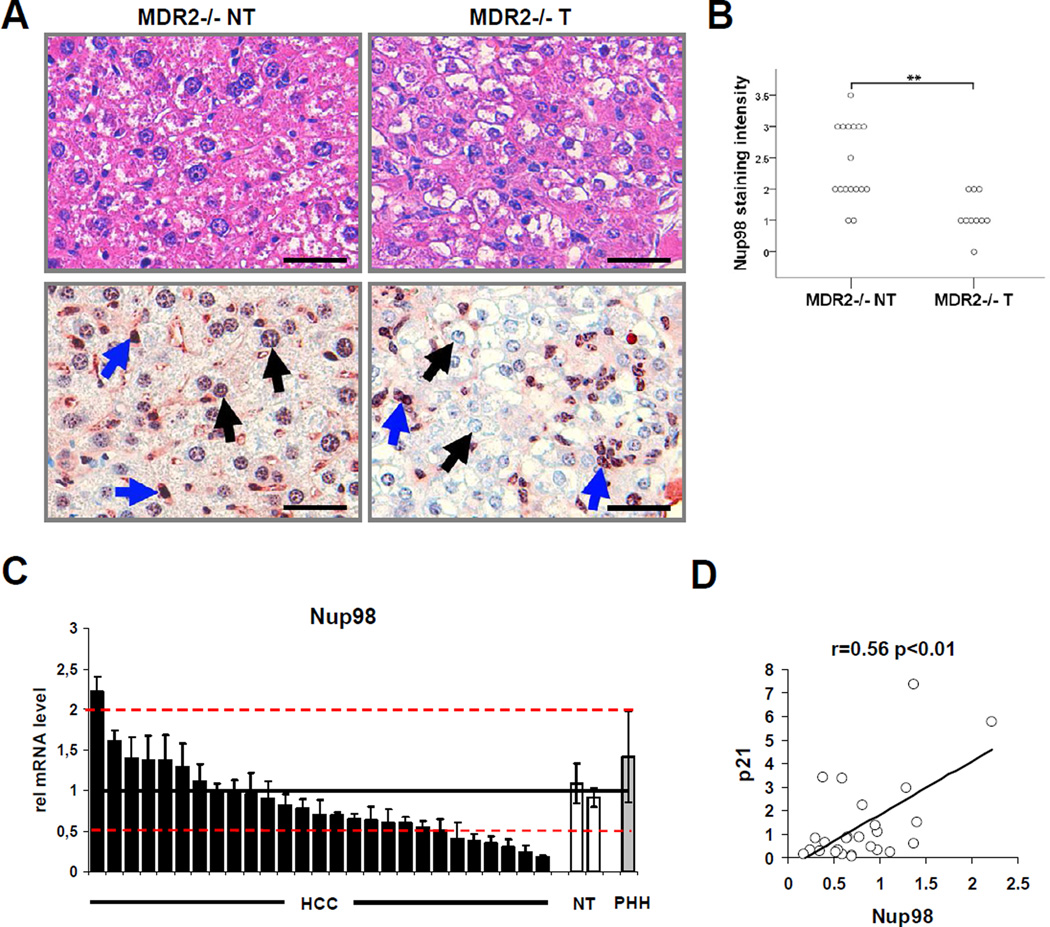
Other than the H1299 lung carcinoma-derived cell line engineered to inducibly express ectopic p21 (H24 p21 cells), the only cell lines in which Nup98 reduction repeatedly produced a strong effect on p21 levels were derived from liver cancers (data not shown) suggesting that Nup98 levels may be altered in hepatocellular carcinoma (HCC). We tested this possibility in both a murine model of HCC and in tissues from HCC patients (Figure 7). The multidrug resistance 2 (MDR-2) knock-out mouse is a model for inflammation-associated HCC development and resembles the human clinical progression of the disease (Mauad et al., 1994). We analyzed Nup98 expression in tumorous (T) and non-tumorous (NT) mouse liver tissue by immunohistochemistry (IHC). Strong Nup98 IHC staining was detected at the nuclear membrane and within the nucleus in hepatocytes (black arrows) and in the sinusoidal cells (blue arrows) of NT-liver tissue (Figure 7A lower left panel). Strikingly, while the sinusoidal cells and the infiltrating inflammatory cells (blue arrows) retained strong Nup98 IHC staining in the T-liver tissue (Figure 7A; lower right panel) there was a significant decrease in Nup98 immunoreactivity (p<0.01) of hepatocellular tumor cells (black arrows) compared to NT-tissue (Figure 7A; lower right panel and see semi-quantitative analysis of hepatocellular Nup98 staining intensity in NT-tissue and T-tissue of MDR-2 −/− mice in Figure 7B). Thus, Nup98 is substantially down-regulated in murine HCC compared to adjacent non-tumorous tissue.
Figure 7. Nup98 Expression Is Downregulated in Hepatocellular Carcinomas (HCCs) in MDR-2 −/− Mice and in Patients with HCC.
(A-B) Lowered hepatocellular Nup98 immunoreactivity in murine tumor tissue compared to non-tumorous tissue.
(A) Corresponding representative liver sections of MDR-2 −/− mice (15 months) either stained with H&E (upper left and upper right panel) or immunohistochemically stained with a polyclonal Nup98 antibody (lower left and lower right panel). Non-tumorous (NT) and tumorous (T) liver tissue is shown in the left and right column respectively. Black arrows indicate nuclei of hepatocytes and hepatocellular tumor cells and blue arrows indicate nuclei of sinusoidal cells and inflammatory cells. Scale bar represents 100 µm.
(B) Diagram shows median values of semiquantiative hepatocellular Nup98 immunohistochemical staining intensities of non-tumorous (NT) and tumorous (T) liver tissue samples of individual MDR-2 −/− mice. Statistical significance was determined from non-parametric testing (Mann-Whitney-U-test, **p<0.01).
(C) Nup98 expression is downregulated in human hepatocellular carcinoma. The relative Nup98 mRNA expression in 27 human hepatocellular carcinoma (HCC) tissue samples (black bars) was assessed by qRT-PCR and compared to the similarly assessed averaged expression (horizontal line) of 2 non-tumorous (NT) liver tissues normalized to 1 (white bars). Red dotted lines indicate thresholds of 2-fold higher or lower expression than the averaged expression in non-tumorous liver tissue. Relative expression in primary hepatocytes (PHH) is also shown (grey bar). 18s-rRNA was used as a housekeeping gene for internal normalization. Data are presented as mean ± SD.
(D) Nup98 and p21 expression are correlated in human hepatocellular carcinoma (HCC). Scatterplot depicts relative Nup98 (x-axis) and p21 (y-axis) mRNA levels measured by qRT-PCR in tumors of the same cohort of HCC patients as in (C). Pearson’s correlation coefficient was used as a measure of association.
To gain insight into a possible role for Nup98 in human hepatocellular carcinoma, we analyzed Nup98 mRNA expression in a cohort of HCC patients comparing HCC tissue samples with NT-liver tissue or with primary hepatocytes (Figure 7C). Nup98 expression was significantly reduced (by a factor of ≥ 2 compared to controls) in 25% of the cases (7/27) and was overexpressed in only 1 out of 27 cases (by ≥ 2-fold compared to control levels). Moreover, Nup98 and p21 expression were associated with a correlation coefficient of r=0.56 (p<0.01) (Figure 7D) suggesting that Nup98-dependent regulation of p21 may also be relevant in vivo. Noteworthy, in a second cohort of HCC patients we found a similar expression pattern with downregulation of Nup98 expression compared to non-tumorous liver in 39% (13/33) and upregulation of Nup98 expression in 12% (4/33) of the cases based on the same thresholds (Figure S4). Again we found a correlation between Nup98 and p21 expression (r=0.40; p<0.05, data not shown). Taken together, these data suggest that Nup98-dependent regulation of p21 is likely to be generalizable to a significant fraction of human HCCs.
Discussion
An RNAi screen revealed that interference with the NPC causes pleiotropic effects on p53 target gene induction. While ablation of a subset of Nups such as Nup98, TPR (translocated promoter region), and Nup93 selectively reduced p21 mRNA accumulation, Aladin knockdown selectively increased p21 mRNA induction. Despite the limitations of this screen (e.g. differences in knockdown efficiency, secondary structural alterations of the NPC upon decreased expression of some Nups, etc) we can assume that selectively reduced p21 mRNA accumulation upon Nup98 ablation is not a general phenomenon observed by depleting any nuclear pore component. Since components of the NPC modulate the expression of p53 target genes in different directions, stabilization of p21 mRNA by Nup98 is unlikely to be the only mechanism that can be envisioned (others include transcriptional regulation, altered protein import/export, etc). This adds another layer of complexity to p53 target gene selection in general and to p21 expression in particular.
Nup98 prevents p21 mRNA degradation by the exosome. This might be related to a role for Nup98 in p21 mRNA export. In fact, a yeast Nup116 deletion mutant exhibits an mRNA export block and accumulation of nuclear mRNAs (Das et al., 2003). Yet, in our single molecule FISH analysis of p21 mRNA, Nup98 knockdown does not lead to increased accumulation of nuclear p21 mRNA. Thus, if knockdown of human Nup98 causes a p21 mRNA export block, this must be tightly coupled to degradation of p21 mRNA transcripts most likely by the nuclear exosome. Indeed, in the Nup116 delta yeast strain, accumulation of nuclear mRNAs is followed by rapid degradation that can be prevented by disruption of the nuclear exosome (Das et al., 2003). In our system, an analogous immediate degradation of unexported p21 mRNA could account for the observed lowered levels of both nuclear and cytoplasmic p21 mRNA. A second scenario can be imagined in which, in addition to its putative effects on p21 mRNA nuclear stability, Nup98 might independently regulate the accumulation of cytoplasmic p21 mRNA. Given the association of Nup98 with the 3’-UTR of p21 mRNA, it may recruit additional factors that stabilize p21 mRNA in the cytoplasm. In either case the exosome likely recognizes and degrades p21mRNA upon Nup98 depletion as a process of mRNA surveillance either related to impaired export or defects in RNA-protein complex formation in the 3’UTR region. Supporting this hypothesis the exosome has been documented to participate in control mechanisms that remove aberrant RNAs in the nucleus and in the cytoplasm (Houseley et al., 2006). Future studies will hopefully elucidate the particular stability defect and subsequent degradation of p21mRNA that occurs in the absence of Nup98.
Bioinformatics analysis identified a C-rich motif that correlates with the stabilizing effect of Nup98 on p21 mRNA and 14-3-3σ mRNA. It will be important eventually to determine which other p53 targets that have high (or low) scores with the C-rich motif are similarly regulated (or not) by Nup98. Particularly relevant to our studies, RNA binding proteins including different members of the poly-C binding protein family (PCBP1, 2 and 4; the latter being a p53 target gene) are implicated in p21 mRNA destabilization via binding to its 3′-UTR (Scoumanne et al., 2010; Waggoner et al., 2009; Zhu and Chen, 2000). Based on the observation that basal p21 mRNA levels are not significantly affected by Nup98 depletion, perhaps Nup98 competes with a p21 mRNA destabilizing factor that is induced by p53, such as MCG-10 (=poly C binding protein 4) (Zhu and Chen, 2000). Alternatively, p21 protein induction may repress expression of a stabilizing factor of its own mRNA. Under basal conditions this factor could compensate for the loss of Nup98. However, under induced conditions (e.g. CPT), this factor would be unavailable to stabilize p21 mRNA due to its depletion via p21-dependent mechanisms. Finally, posttranslational modifications of Nup98 upon p53/p21 activation may be required for the stabilizing effect on the p21mRNA transcript. Exploring aspects of these proposed scenarios will be of considerable interest for further studies.
Data from the murine HCC model, combined with data from both cohorts of HCC patients (Figure 7 and S4) suggest that it may be advantageous for tumors to lose Nup98 expression i.e. that Nup98 acts under certain circumstances as a tumor suppressor in hepatocarcinogenesis. In line with this is the observation that p53-mediated induction of senescence, an important liver tumor-suppressive response (Xue et al., 2007), is impaired upon Nup98 depletion (Figure 6). However, more studies including mouse models with a liver-specific conditional Nup98 knock-out are required to draw firm conclusions about tumor-suppressive functions of Nup98 that might be context- and stress-specific. In fact, since up to 12% of HCC patients showed overexpression of Nup98 it is quite possible that Nup98 may play a dual role in cancer formation as does p21 or, even more intriguingly, through p21. The latter could be supported by the correlation of Nup98 and p21 expression in both patient data sets. We expect that future studies will elucidate the specific circumstances under which Nup98 may act as barrier to cancer formation.
Experimental Procedures
Cell Culture and Drug Treatment
HepG2 cells were maintained in RPMI medium supplemented with 10% fetal bovine serum (FBS). Hep3B cells and the derivatives Hep3B-4Bv (expressing the temperature sensitive p53val135 mutation) were kindly provided by M. Oren (Friedman et al., 1997) and maintained in MEM with 10% FBS (and 1 µg/mL puromycin for Hep3B-4Bv). H1299 cells and their derivative ‘tet-off’ H24-p21 inducible cell line were previously described (Niculescu et al., 1998) and maintained in DME medium with 10% FBS and tetracycline (5 µg/ml). p21 expression was induced by removing tetracycline 48 h before harvesting the cells. H1299 PUMA-inducible cells (‘tet-on’) were generously provided by K. Vousden (Nakano and Vousden, 2001) and grown in DME with 10% FBS. PUMA expression was induced by adding Doxycycline (2 µg/mL) 24 h before harvesting. Camptothecin (CPT), Nutlin-3a, and Actinomycin D (Sigma) were used as indicated.
Transfections and Plasmids
siRNA transfection assays were performed with DharmaFECT 1 (Dharmacon) or Oligofectamin (Invitrogen). siRNA duplexes, designed and synthesized by Qiagen were used individually at 50 nM (sequences are listed in Table S3). The Qiagen All-Stars duplex was used as the negative control siRNA for all knockdown experiments besides the p21 mRNA half-life experiment, where Luc siRNA was used. HA-Nup98 was generously provided by Dr. N.R. Yaseen (Washington University School of Medicine, USA). GFP-Nup98 full length, GFP-Nup98 C-terminus (506–920), GFP-Nup98 GLFG+C-terminus (221–920), GFP-Nup98 GLFG(C)+C-terminus (362–920) were as described (Griffis et al., 2004).. HA-Nup96 was kindly provided by Dr. B. Fontoura (UT Southwestern Medical Center, USA). cDNAs encoding p21 full length (FL), PUMA full length (Pu FL) including the respective 5’ and 3’ UTRs and p21 lacking the 3’-UTR were amplified by PCR with a Platinum Pfx DNA polymerase (Invitrogen) using cDNA from HepG2 cells as template. The resulting p21 FL and p21 delta 3’-UTR cDNAs were then cloned into a pcDNA3.1 vector (Invitrogen). The Pu/p21 hybrid construct was generated by amplifying the PUMA CDS (including the 5’UTR) and p21 3’UTR from the respective FL constructs, that were sequentially cloned into a pcDNA3.1 vector (Invitrogen). The Luc/p21 3’UTR and Luc control were as described (Giles et al., 2003). Transient plasmid transfections were done with Lipofectamin2000 (Invitrogen) according to the manufacturer’s protocol.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
RNA was isolated from cells with the Qiagen RNeasy Mini kit, and was DNAse treated and reverse transcribed using the Qiagen Quantitect reverse transcription kit. Samples were analyzed on an ABI 7300 real-time PCR instrument with default settings using the ΔΔCT method. Expression levels were normalized to those of RPL32 or 18s RNA (patient samples). Primers used were designed with Primer Express (Applied Biosystems) and sequences are listed in Table S4.
Immunoblots and Antibodies
Cells were harvested and subjected to immunoblotting as previously described (Ohkubo et al., 2006). Commercial antibodies used in the study are listed in table S5. Nup98 rabbit polyclonal antibody was kindly provided by Dr. G. Blobel (Rockefeller University, USA) and Nup96 rabbit polyclonal antibody was a kind gift from Dr. Joseph Glavy (Stevens Institute of Technology, USA). Signal detection was performed by the using the Odyssey Infrared Imaging System (LI-COR).
Immunofluorescent Staining
HepG2 and H24 p21 cells were grown on coverslips and treated with drugs as indicated. Immunofluorescent staining was performed as previously described (Karni-Schmidt et al., 2007). Nup98 (C-5; Santa-Cruz) was used as primary antibody followed by Alexa Fluor 488 rabbit anti-mouse secondary antibody (Molecular Probes, Eugene, OR). Images were analyzed by confocal laser scanning microscopy (Olympus model 1381) using Fluoview software (Canter Valley, PA).
Fluorescence-Activated Cell Sorting (FACS)
Cells were processed with slight modifications as described before (Ohkubo et al., 2006). Cell cycle stages were analyzed by using the ModFit LT version 3.0 program and subG1 fractions were analyzed using Cell Quest Pro.
Fluorescence In Situ Hybridization (FISH)
To detect p21 mRNA 14 oligodeoxynucleotide probes were designed, synthesized and labeled as previously described (Femino et al., 1998). Each probe had a length of 50 nt and contained five amino-modified nucleotides (amino-allyl T). The free amines were chemically coupled to cyanine 5 fluorescent dye after synthesis. The sequences of probes used to detect p21 mRNA are available upon request. mRNA-FISH was performed based on a published protocol (http://www.singerlab.org/protocols). For further information see the Supplemental Experimental Procedures.
RNA Immunoprecipitation
RNA-IP was performed using a published protocol (Kaneko and Manley, 2005) with slight modifications. Further information is provided in the Supplemental Experimental Procedures.
Animals and Immunohistochemical Staining
MDR-2 −/− animals were described previously (Mauad et al., 1994). Liver tissue of 15 month old male MDR-2 −/− mice was immunostained based on a published protocol (Singer et al., 2007) by using a polyclonal Nup98 antibody. For additional information see the Supplemental Experimental Procedures.
Patient Data
Total RNA isolation of HCC samples (n=27 and n=33) and healthy liver tissues (n=2 and n=5) of both cohorts for semiquantitative real-time PCR was performed using the NucleoSpin RNA II kit according to the manufacturers' protocol (Macherey-Nagel, Duren, Germany). The study was approved by the institutional ethics committee of the Medical Faculty at Heidelberg University.
Motif Discovery
We used the MatrixREDUCE program from the REDUCE software suite (http://bussemakerlab.org/software/REDUCE) to perform a genomewide fit of a position-specific affinity matrix (PSAM) to the logarithm of mRNA expression fold-differences between wild-type cells and p53 knock-down cells (after 72 h). For the sequence associated with each log-ratio, we used the March 2006 version of human 3′-UTRs from the UCSC genome website (http://genome.ucsc.edu). Genes with overlapping 3′-UTRs were filtered out. In the PSAM search, we considered motif lengths from 1 to 10 and used a p-value threshold of 0.001. To rank the p53 target genes, we calculated the total affinity of the 3′-UTR using the AffinityProfile program from the REDUCE suite to score the enrichment of each 3′-UTR for the discovered C-rich motif. Total relative affinities were calculated as the sum of PSAM-based predicted affinities in a sliding window over the length of the 3′-UTR. We controlled for variation in 3′-UTR length by dividing the total affinity by the UTR length and used the resulting affinity density to rank the list of published p53 targets (Riley et al., 2008).
Supplementary Material
01
Highlights.
- Nup98 is required for full induction of select p53 target genes such as p21
- Nup98 associates with the 3’-UTR of p21 mRNA preventing exosomal degradation
- Nup98 is downregulated in murine and human HCC and correlates with p21 expression
Acknowledgements
We thank Ella Freulich, Melissa Lopez-Jones, Juliane Winkler, Helen Mueller, Michaela Bissinger, Andrea Hain and Eva Eiteneuer for excellent technical assistance, Oleg Laptenko, Will Freed-Pastor and other members of the Prives Laboratory for helpful suggestions, Gabor Halasz for compiling a non-redundant set of 3′-UTR sequences, and Xiang-Jun Lu for implementing and maintaining the REDUCE software suite. S.S. was supported by a fellowship from the German Research Foundation Si-1487/1-1 and SFB/TRR77-C5. J.H. and T.P. were supported by SFB/TRR-A7, K.B. by SFB/TRR-B7, T.L. and O.N. by SFB/TRR-B5. This research was supported by NIH grant CA77742 to C.P. DG and the imaging services were partially supported by NIH GM86217 to RHS. HJB was supported by NIH grants R01HG003008 and U54CA121852, as well as a John Simon Guggenheim Foundation Fellowship. MF was supported by NIH training grant T32GM082797.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem. 2003;278:20979–20988. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau BN, Diaz RL, Saunders MA, Cheng C, Chang AN, Warrener P, Bradshaw J, Linsley PS, Cleary MA. Identification of SULF2 as a novel transcriptional target of p53 by use of integrated genomic analyses. Cancer Res. 2009;69:1368–1374. doi: 10.1158/0008-5472.CAN-08-2742. [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Butler JS, Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enninga J, Levy DE, Blobel G, Fontoura BM. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science. 2002;295:1523–1525. doi: 10.1126/science.1067861. [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc Natl Acad Sci U S A. 2005;102:17675–17680. doi: 10.1073/pnas.0503803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL, Shaulian E, Littlewood T, Resnitzky D, Oren M. Resistance to p53-mediated growth arrest and apoptosis in Hep 3B hepatoma cells. Oncogene. 1997;15:63–70. doi: 10.1038/sj.onc.1201149. [DOI] [PubMed] [Google Scholar]
- Giles KM, Daly JM, Beveridge DJ, Thomson AM, Voon DC, Furneaux HM, Jazayeri JA, Leedman PJ. The 3'-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J Biol Chem. 2003;278:2937–2946. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Ezra O, Rivlin N, Molchadsky A, Madar S, Goldfinger N, Rotter V. p53, a novel regulator of lipid metabolism pathways. J Hepatol. 2012;56:656–662. doi: 10.1016/j.jhep.2011.08.022. [DOI] [PubMed] [Google Scholar]
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcriptiondependent mobility. Mol Biol Cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Manley JL. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3' end formation. Mol Cell. 2005;20:91–103. doi: 10.1016/j.molcel.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Karni-Schmidt O, Friedler A, Zupnick A, McKinney K, Mattia M, Beckerman R, Bouvet P, Sheetz M, Fersht A, Prives C. Energy-dependent nucleolar localization of p53 in vitro requires two discrete regions within the p53 carboxyl terminus. Oncogene. 2007;26:3878–3891. doi: 10.1038/sj.onc.1210162. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Mauad TH, van Nieuwkerk CM, Dingemans KP, Smit JJ, Schinkel AH, Notenboom RG, van den Bergh Weerman MA, Verkruisen RP, Groen AK, Oude Elferink RP, et al. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am J Pathol. 1994;145:1237–1245. [PMC free article] [PubMed] [Google Scholar]
- Moore MA, Chung KY, Plasilova M, Schuringa JJ, Shieh JH, Zhou P, Morrone G. NUP98 dysregulation in myeloid leukemogenesis. Ann N Y Acad Sci. 2007;1106:114–142. doi: 10.1196/annals.1392.019. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S, Tanaka T, Taya Y, Kitazato K, Prives C. Excess HDM2 impacts cell cycle and apoptosis and has a selective effect on p53-dependent transcription. J Biol Chem. 2006;281:16943–16950. doi: 10.1074/jbc.M601388200. [DOI] [PubMed] [Google Scholar]
- Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Rosenblum JS, Blobel G. Autoproteolysis in nucleoporin biogenesis. Proc Natl Acad Sci U S A. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoumanne A, Cho SJ, Zhang J, Chen X. The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S, Ehemann V, Brauckhoff A, Keith M, Vreden S, Schirmacher P, Breuhahn K. Protumorigenic overexpression of stathmin/Op18 by gain-of-function mutation in p53 in human hepatocarcinogenesis. Hepatology. 2007;46:759–768. doi: 10.1002/hep.21736. [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Waggoner SA, Johannes GJ, Liebhaber SA. Depletion of the poly(C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J Biol Chem. 2009;284:9039–9049. doi: 10.1074/jbc.M806986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Powers MA. Nuclear pore proteins and cancer. Semin Cell Dev Biol. 2009;20:620–630. doi: 10.1016/j.semcdb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen X. MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G(2)-M. Mol Cell Biol. 2000;20:5602–5618. doi: 10.1128/mcb.20.15.5602-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01