G Protein–Coupled Receptor–Mediated Activation of p110β by Gβγ Is Required for Cellular Transformation and Invasiveness (original) (raw)
. Author manuscript; available in PMC: 2014 Apr 8.
Published in final edited form as: Sci Signal. 2012 Dec 4;5(253):ra89. doi: 10.1126/scisignal.2003264
Abstract
Synergistic activation by heterotrimeric guanine nucleotide–binding protein (G protein)-coupled receptors (GPCRs) and receptor tyrosine kinases distinguishes p110β from other class IA phosphoinositide 3-kinases (PI3Ks). Activation of p110β is specifically implicated in various physiological and pathophysiological processes, such as the growth of tumors deficient in phosphatase and tensin homolog deleted from chromosome 10 (PTEN). To determine the specific contribution of GPCR signaling to p110β-dependent functions, we identified the site in p110β that binds to the Gβγ subunit of G proteins. Mutation of this site eliminated Gβγ-dependent activation of PI3Kβ (a dimer of p110β and the p85 regulatory subunit) in vitro and in cells, without affecting basal activity or phosphotyrosine peptide–mediated activation. Disrupting the p110β-Gβγ interaction by mutation or with a cell-permeable peptide inhibitor blocked the transforming capacity of PI3Kβ in fibroblasts, and reduced proliferation, chemotaxis, and invasiveness of PTEN-null tumor cells in culture. Our data suggest that specifically targeting GPCR signaling to PI3Kβ could provide a therapeutic approach for tumors that depend on p110β for growth and metastasis.
Introduction
Signaling by class I phosphoinositide 3-kinases (PI3Ks) is commonly enhanced in tumors by gene amplification, activating mutations, or the inactivation of phosphatase and tensin homolog deleted from chromosome 10 (PTEN), a tumor suppressor lipid phosphatase [1]. Class I PI3Ks produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) in cells and stimulate proliferation, survival, and motility. The class IA enzymes are obligate heterodimers consisting of distinct catalytic (p110) subunits bound to the same regulatory (p85) subunits [2, 3]. Among the three class IA PI3Ks, the PIK3CB gene product p110β is unique, because it can be activated both by receptor tyrosine kinases (RTKs) and downstream of heterotrimeric guanine nucleotide-binding protein (G protein)–coupled receptors (GPCRs) through direct binding to Gβγ subunits [4–7]. PTEN-deficient prostate cancer development specifically depends on the activity of the p110β-p85 dimer (referred to as PI3 Kβ), but the mechanism for this specificity is currently unknown [8–11]. Whether GPCRs have a role in PI3Kβ-mediated transformation of PTEN-null cells has remained an open question because of the lack of tools to specifically probe the Gβγ-PI3Kβ interaction.
Defining the role of Gβγ in activating effectors such as p110β is challenging because of the transient nature of interactions between the two and because of the lack of a distinct Gβγ-binding motif that could be used to identify its target binding sites. This contrasts with the mechanism of activation of PI3Ks by RTKs, which involves high-affinity interactions that have been well characterized [12, 13]. To investigate the mechanism of p110β activation downstream of GPCRs by Gβγ, and to define the role of this interaction in p110β signaling in cells, we have identified the Gβγ-binding site on p110β. We took two parallel approaches, the first based on an analysis of sequence conservation, and the second with hydrogen-deuterium exchange mass spectrometry (HDX-MS). Both approaches identified the same region, enabling us to generate a p110β mutant that remained sensitive to stimulation by RTKs but did not respond to activation by Gβγ. This mutant enabled us to interrogate the physiological importance of p110β activation downstream of GPCRs by Gβγ, and to define a critical role for this interaction in the cellular transformation, proliferation, and chemotaxis of PTEN -null tumor cells.
Results
Identification of the Gβγ-binding site in p110β
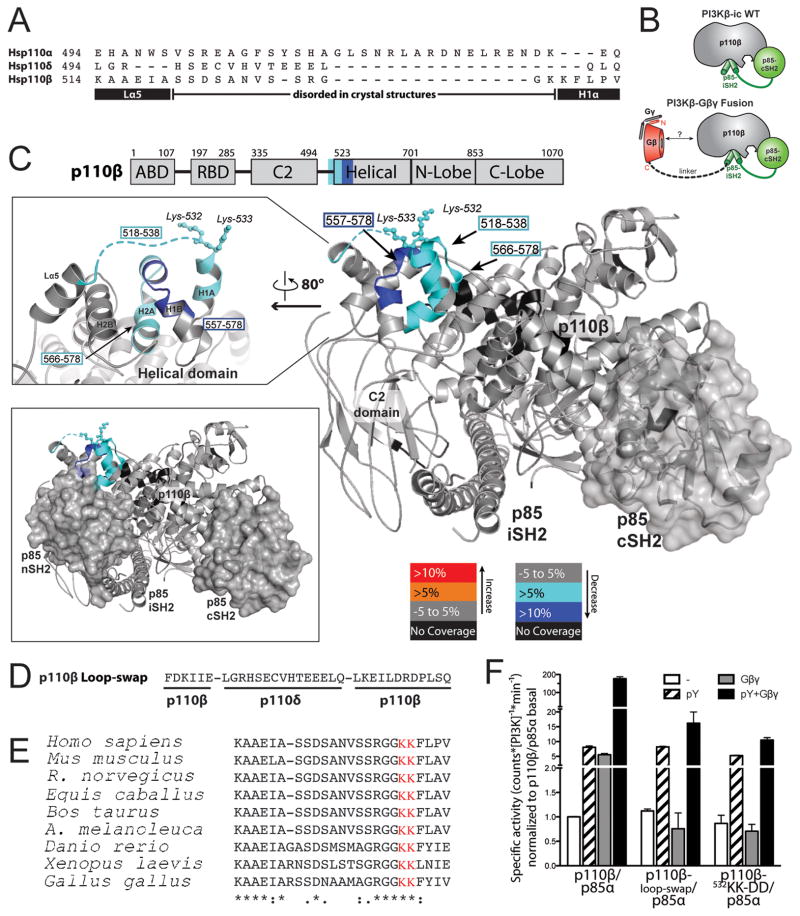
We previously showed that the adaptor-binding, Ras-binding, and C2 domains of p110β are not responsible for its activation by Gβγ subunits [14]. For this reason, we compared the remainder of the p110β sequence with those of p110α and p110δ, which are insensitive to stimulation by Gβγ, to look for sequence differences that might account for the selective activation of p110β by Gβγ. Whereas the helical and kinase domains of all three isoforms display high sequence similarity, we identified a 24-amino acid residue, non-conserved region (residues 514 to 537) in the linker between the C2 domain and the helical domain of p110β (Fig. 1A and fig. S1). The central portion of this segment is not visible in the crystal structure of p110β, presumably because it is disordered, but it is part of a surface-accessible loop [15].
Fig. 1.
Mapping of the Gβγ-binding site on p110β by sequence analysis and HDX-MS. (A) Sequence alignment of the C2 domain–helical domain linker region of p110α, β, and δ. The black rectangles denote helices in the p110β structure, and the black line represents the disordered region. (B) Cartoon illustration of the p110β–p85α-icSH2 wild-type (WT) heterodimer and the p110β–Gβ-p85α-icSH2–Gγ-C68S fusion heterotrimer (fusion) used for the HDX-MS experiments. (C) Domains of p110β are outlined and colored according to the legend for changes associated with the presence of Gβγ. Regions in p110β and p85α-icSH2 that showed >0.5 Dalton and >5% changes in deuteration extent between the WT and fusion complexes were mapped on the p110β–p85β–icSH2 model (PDB: 2y3a, right panel). The loop region between the C2 domain and the helical domain is represented as a dotted line because it is not ordered in the structure. Residues corresponding to human p110β K532 and K533 are represented with balls and sticks. Top left, a close-up view of the p110β region in which changes in deuteration extent as a result of the presence of Gβγ were detected is shown. Bottom left, a model for the p110β–p85α-nicSH2 generated by combining the structures of p110β–p85β-icSH2 (PDB: 2Y3A) and p85α–nSH2 (PDB: 3HHM). The nSH2 and cSH2 domains of p85 are shown as surface representations. The p85α-nSH2 position is based on the structure of p110α, although there is no unambiguous evidence that nSH2 adopts exactly the same position when in complex with p110β. (D) Sequence of the loop-swap mutant of p110β. (E) Alignment of p110β zoologs in the region of the C2-helical linker. (F) Activities of WT PI3Kβ and the loop-swap and 532KK-DD mutants purified from insect cells, in the presence of pY peptide (pY) and lipidated Gβγ. Activities were expressed relative to the basal activity of PI3Kβ, which was normalized to one. Graph shows the activity ±SD of three independent experiments.
In parallel, we used an empirical approach, HDX-MS, to experimentally identify the p110β-Gβγ interaction sites. HDX-MS is a powerful technique to monitor protein dynamics, protein-protein interactions, as well as protein-lipid interactions [16–19]. For HDX-MS measurements, we used two experimental setups, one with soluble Gβγ (Gγ-C68S) (Fig. 1, B and C), and another with lipid-modified Gβγ in the presence of membranes (fig. S2). To enhance the stability of interaction between the p110β-p85 dimer and soluble Gβγ in solution, we produced a heterotrimer containing p110β, Gγ C68S, and a chimeric construct containing Gβ covalently linked to a fragment of p85α containing the C-terminal Src homology 2 (SH2) domain and the coiled-coil domain (iSH2-cSH2) (Fig. 1B). This heterotrimer formed a stable complex that could be stimulated by both a platelet-derived growth factor receptor (PDGFR)-derived bis-phosphopeptide (pY) and Gβ1γ2 subunits (Gβγ) (fig. S3A). When we compared differences in the hydrogen-deuterium (HD) exchange rates of p110β peptides between the heterotrimeric fusion complex and the wild-type p110β-p85α-icSH2 heterodimer, we identified two stretches that were more protected in the fusion complex (Fig. 1C and fig. S3, B and C). The first potential Gβγ-binding site, containing residues 518 to 538, matched very well with the region mapped by sequence analysis. The second protected region, amino acids 557 to 578, lies underneath the C2-helical linker. Changes in this region are likely a result of indirect effects from the binding of Gβγ to the linker above it. The same regions of p110β binding to Gβγ were identified with full-length PI3Kβ and lipidated Gβγ, with liposomes to stabilize the interactions (fig. S2, C and D, tables S1 to S6). Taken together, the sequence analysis and HDX-MS data suggested that the region of p110β spanning residues 518 to 537 was the Gβγ-binding site.
To test whether this region was involved in Gβγ-mediated regulation of p110β, we designed a loop-swap p110β mutant in which these 24 amino acid residues were replaced with the corresponding region of p110δ (Fig. 1D). We also mutated residues lysines 532 and 533 (532KK) in the p110β loop, which are highly conserved among p110β from different species but not between p110β and p110α or p110δ, and which are ordered in the p110β crystal structure (Fig. 1E) [20]. Replacement of the p110β loop with that of p110δ or mutation of 532KK to DD had no effect on the in vitro basal kinase activity of PI3Kβ in assays with purified enzyme from insect cells (Fig. 1F) or in assays with enzyme immunopurified from mammalian cells (fig. S4, A and B). However, whereas wild-type PI3Kβ was markedly activated by the addition of Gβγ, neither the 532KK-DD mutant nor the loop-swap mutant of PI3Kβ was activated by Gβγ (Fig. 1F, figs. S4 A and B). Wild-type and mutant enzymes were activated to a similar extent by pY (Fig. 1F, S4A, S4B), even though the mutation sits close to the predicted p85-nSH2–binding site (Fig. 1C) [21]. Similar results were obtained with a 514KAAEI-DAAKA mutant of p110β, which targets the N-terminal end of the loop (fig. S4C). The degree of activation of PI3Kβ by pY and Gβγ (Fig. 1) was consistent with previous studies with baculovirally-expressed PI3Kβ purified from insect cells [7, 15], although it was substantially greater than that seen with PI3Kβ immunopurified from transfected mammalian cells (fig. S4). These differences in fold activation may reflect the influences of assay conditions (see the Supplementary Materials), N-terminal tags on basal activity, or the presence of an antibody bound to the immunopurified enzyme [2, 22].
Role of Gβγ-mediated activation of p110β in signaling, transformation, and cell motility
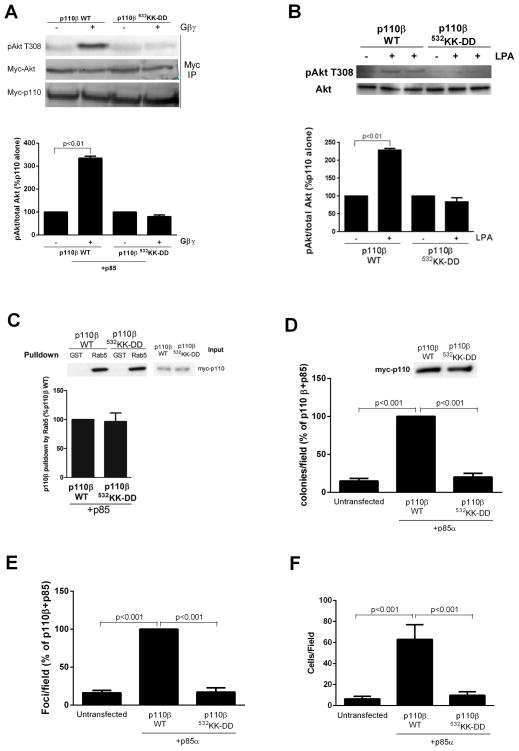
To measure the effect of the p110β mutation on signaling to the serine and threonine kinase Akt, we transfected human embryonic kidney (HEK) 293Ecells with plasmids encoding the wild-type or 532KK-DD mutant p110β together with plasmids encoding p85α and myc-tagged Akt (myc-Akt), with or without plasmids encoding Gβ1γ2 subunits, which activate p110β in vitro [23]. Gβγ-dependent Akt activation in this system was specifically inhibited by the p110β inhibitor TGX-221, and therefore reflected Gβγ-mediated stimulation of p110β (fig. S5A). Whereas cells containing wild-type PI3Kβ showed a marked increase in the abundance of Akt activated by phosphorylation at Thr308 (pT308-Akt) in the presence of exogenous Gβγ subunits, cells transfected with plasmid encoding the 532KK-DD mutant PI3Kβ showed a complete loss of Akt activation in the presence of Gβγ subunits (Fig. 2A). Similarly, lysophosphatidic acid (LPA)-stimulated Akt activation was greater in NIH3T3 cells stably expressing wild-type PI3Kβ than in cells expressing the 532KK-DD mutant p100β (Fig. 2B). The 532KK-DD mutation had no effect on the binding of p110β to the small guanosine triphosphatase (GTPase) Rab5 (Fig. 2C), indicating that interactions of mutant p110β with other intracellular regulators were intact. These data show that the C2-helical linker region of p110β is necessary for Gβγ-mediated activation of PI3Kβ in vitro and in cells.
Fig. 2.
Role of Gβγ in PI3Kβ-mediated signaling, transformation, motility, and invasion. (A) HEK 293E cells were transfected with plasmids encoding myc-Akt and either WT or the 532KK-DD mutant PI3Kβ, with or without plasmids encoding Gβγ. Akt activation in samples immunoprecipitated (IP) with an antibody against myc was analyzed by Western blotting with an antibody against pT308-Akt. The ratio of the amount of pAkt to that of total Akt is expressed as a percentage of that under basal conditions. (B) NIH3T3 cells stably expressing WT or mutant PI3Kβ were stimulated with 10 nM LPA for 5 min. Akt activation was analyzed by Western blotting with anti-pT308-Akt antibody and quantified as described earlier. (C) HEK 293T cells were transfected with plasmid encoding WT or 532KK-DD mutant p110β. Cell lysates were incubated with GST or GST-Rab5 immobilized on glutathione-Sepharose beads, and bound material was analyzed by Western blotting. Graphs in each panel show the mean percentage pulldown ± SEM from three separate experiments. (D and E) NIH3T3 cells were transfected with plasmids encoding p85α and either WT or the 532KK-DD mutant p110β, and (D) the formation of colonies in soft agar or (E) the formation of foci were measured. Graphs in each panel show means ± SEM from three separate experiments. (F) Migration of control NIH3T3 cells or cells stably expressing WT or mutant PI3Kβ towards FBS was measured in a Boyden chamber assay. Assays were conducted in triplicate, and the data are pooled from two separate experiments.
To test the biological relevance of Gβγ-mediated activation in p110β signaling, we compared the ability of wild-type and mutant p110β constructs to mediate cellular transformation and motility. In a soft agar colony formation assay, cells transfected with plasmid encoding wild-type PI3Kβ generated substantially more colonies than did control cells. However, transfection of cells with plasmid encoding the 532KK-DD mutant PI3Kβ resulted in a complete loss of transformation (Fig. 2D, fig. S5B). Similar results were obtained in a focus formation assay, in which NIH3T3 cells transfected with plasmid encoding wild-type PI3Kβ formed a substantially greater number of foci compared to that formed by cells transfected with plasmid encoding the 532KK-DD mutant PI3Kβ (Fig. 2E, fig. S5C). This result was not because of differences in proliferation, because wild-type and mutant p110β caused similar increases in proliferation as compared to that of control NIH 3T3 cells (fig. S6A). Similarly, wild-type, but not mutant, p110β increased the extent of chemotaxis of cells toward serum in a Boyden chamber assay (Fig. 2F). The ability of wild-type p110β to enhance transformation and migration was Gβγ-dependent, because it was inhibited by pertussis toxin (fig. S6, B and C). These data showed that GPCR inputs to PI3Kβ, transmitted by Gβγ, are critical for PI3Kβ-mediated cellular transformation and enhancement of motility.
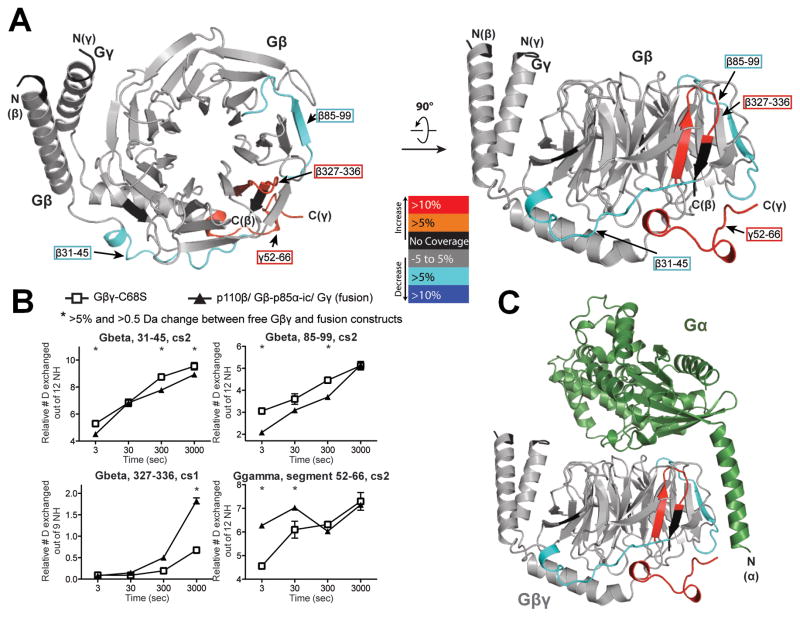
Identification of the p110β-binding site on Gβγ by HDX-MS
To explore the possibility of specifically inhibiting the interactions between p110β and Gβγ, we used the HDX-MS approach to determine the region on Gβγ that binds to p110β (Fig. 3). We identified two regions on Gβγ that were more protected in the fusion compared to in the free Gβγ. One of the peptides spans residues 31 to 45 in Gβ, in the linker between the N-terminal α-helix and the first blade of the β-propeller (Fig. 3, A and B). This region was not previously observed to interact with other Gβγ effectors. The other more protected stretch spans residues 85 to 99 in the second blade of the β-propeller, a region previously identified to be of major importance for the activation of phospholipase C β2 (PLC-β2) [24, 25]. This region contains the residue Trp99 at the top of the propeller, which is part of the “hot-spot” region in Gβ that makes contacts with several effectors [26]. These data showed that p110β shares a common Gβγ-binding surface with other effectors, such as the G protein α-subunit (Fig. 3C), PLC-β, adenylyl cyclase [27], and PI3Kγ [28], but that it also uniquely affects a linker region between the N-terminal α-helix and the first blade of Gβ. This interface could provide an attractive target for therapeutics, because targeted disruption of this interface should have relatively specific effects on Gβγ-mediated activation of p110β.
Fig. 3.
Mapping of the p110β-binding region in Gβγ heterodimers with HDX-MS. (A) The p110β–Gβ-p85α-icSH2–Gγ-C68S fusion heterotrimer (fusion) was used to compare deuterium incorporation with that of free Gβγ-C68S (Gβγ). Regions in Gβ and Gγ that showed >0.5 Dalton and >5% changes between free Gβγ and the fusion were mapped onto the Gβγ model (PDB ID: 1GOT). In addition to the protected peptides described in the text, there was some exposure of the C-terminus of Gβ and the adjacent C-terminus of Gγ, which were probably a consequence of the attachment of the C-terminus of Gβ to the linker connecting to p85 in the fusion. (B) All peptides in Gβ and Gγ that showed changes in deuteration extent between free Gβγ and the fusion proteins are shown. The stretch of amino acid residues 52 to 66 in Gγ is labeled as a segment to denote that these data were generated by subtraction of the deuterium incorporation of peptide 44 to 51 from that of peptide 44 to 66. * indicates changes that were >0.5 Dalton and >5%. Experiments were performed in duplicate and graphs show the SD. (C) Crystal structure of Gβγ bound to Gα (PDB ID:1GOT). Gβγ is colored as in (A).
Inhibition of the proliferation, chemotaxis and invasion of PTEN-null tumor cells by a peptide inhibitor of the p110β-Gβγ interaction
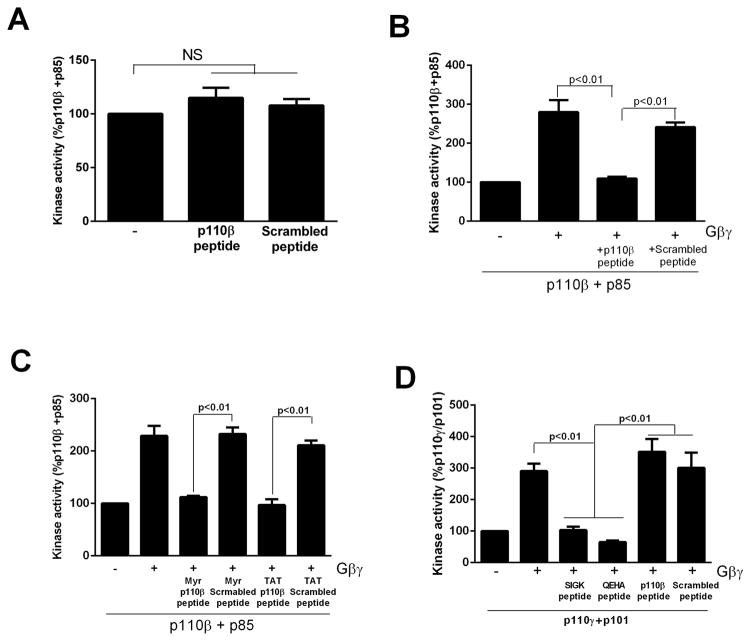
To generate an inhibitor of p110β-Gβγ interactions, we synthesized a peptide derived from the C2-helical linker region of p110β (514KAAEIASSDSANVSSRGGKKFLPV). The peptide had no effect on basal PI3Kβ activity (Fig. 4A), but blocked Gβγ-dependent activation of PI3Kβ in vitro, whereas a scrambled peptide had no effect (Fig. 4B). Similar results were obtained with N-myristoylated and N-TAT-labeled versions of the peptide (Fig. 4C), which are cell-permeable versions of the peptide. The peptide had no effect on Gβγ-mediated activation of p101–p110γ dimers, which were inhibited by peptides that target the canonical Gβγ effector–binding site (SIGK and QEHA, Fig. 4D).
Fig. 4.
A peptide derived from p110β blocks the activation of PI3Kβ by G βγ in vitro. (A). PI3Kβ immunopurified from HEK 293T cells was incubated in the absence or presence of 1 μM p110β-peptide or scrambled peptide and assayed for lipid kinase activity. (B) PI3Kβ immunopurified from HEK 293T cells was incubated in the absence or presence of recombinant lipidated Gβγ and 1 μM p110β-peptide or scrambled peptide and assayed for lipid kinase activity. (C) PI3Kβ immunopurified from HEK 293T cells was incubated in the absence or presence of recombinant lipidated Gβγ and 1 μM myristoylated or TAT-tagged p110β-peptide or scrambled peptide and assayed for lipid kinase activity. (D) Immunopurified p101–p110γ from HEK 293T cells was incubated with or without recombinant lipidated Gβγ and 1 μM p110β-peptide, scrambled peptide, 1 μM QEHA peptide, or 10 μM SIGK peptide. Data in all panels are the means ± SEM of triplicate measurements and are representative of two to three experiments.
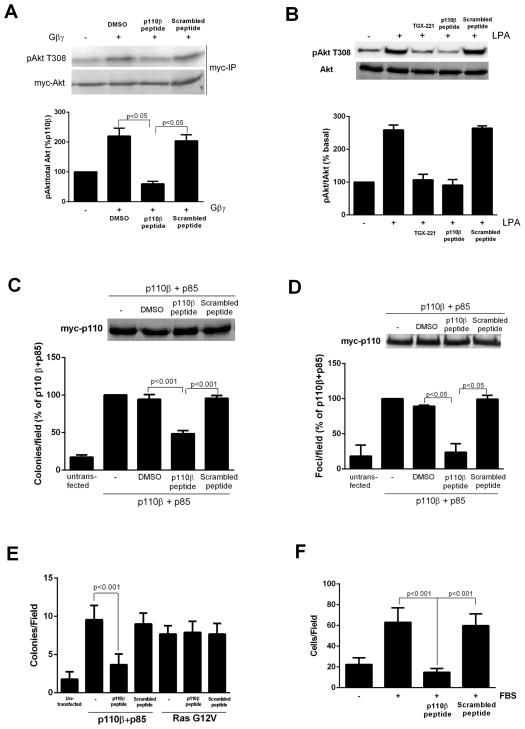
We tested the effects of the myristoylated peptide on Gβγ-mediated activation of Akt in NIH3T3 cells transfected with plasmid encoding myc-Akt with or without plasmids encoding Gβγ subunits. The myristoylated p110β-peptide completely inhibited Gβγ-mediated (Fig. 5A) and LPA-stimulated (Fig. 5B) increases in Akt phosphorylation at Thr308 at a concentration of 30 μM (fig. S7A), whereas myristoylated scrambled peptide or vehicle control had minimal effects. In contrast, the myristoylated peptide had no effect on epidermal growth factor (EGF)-dependent activation of Akt in a cell line in which TGX221 had a substantial inhibitory effect (fig. S7B), which showed that the myristoylated peptide had no effect on RTK-mediated activation of PI3Kβ in intact cells. The inhibitory activity of the myristoylated peptide required its entry into the cells, because both myristoylated and TAT-tagged peptides inhibited Gβγ-dependent activation of Akt, whereas unlabeled peptide had no such effect (fig. S7C). In addition, the myristoylated p110β-peptide, but not the myristoylated scrambled peptide, inhibited PI3Kβ-dependent transformation of NIH3T3 cells, as was observed in a soft agar colony formation assays (Fig. 5C and fig. S5B) and focus formation assays (Fig. 5D and fig. S5C). Inhibition of cellular transformation by the myristoylated p110β-peptide was not a result of decreased proliferation, because neither the myristoylated peptide nor pertussis toxin inhibited the proliferation of NIH3T3 cells transfected with plasmid encoding PI3Kβ (fig. S7D). In contrast, the myristoylated peptide had no effect on cellular transformation caused by expression of oncogenic Ras (Fig. 5E). The myristoylated peptide also blocked the enhanced migration in a Boyden chamber assay of NIH3T3 cells transfected with plasmid encoding PI3Kβ(F ig.5F).
Fig. 5.
Peptide inhibitors disrupt PI3Kβ activation and signaling in response to Gβγ. (A) HEK 293E cells were transfected with plasmids encoding p110β, p85, and myc-Akt with or without plasmid encoding Gβγ. Cells were treated with 30 μM peptide or scrambled peptide for 30 min, and the extent of phosphorylation of Akt at Thr308 (T308) was determined by Western blotting analysis. (B) NIH3T3 cells were pretreated with TGX221, p110β-peptide, or scrambled peptide and stimulated with 10 nM LPA for 5 min before the extent of phosphorylation of Akt at Thr308 was determined by Western blotting analysis. (C) NIH3T3 cells were transfected with plasmids encoding WTp110β and p85α, and colony formation in soft agar was measured in the absence or presence of 30 μM p110β-derived myristoylated peptide or scrambled peptide. (D) NIH3T3 cells were transfected with plasmids encoding WT p110β and p85α, and foci formation was measured in the absence or presence of 30 μM p110β-derived myristoylated peptide or scrambled peptide. (E) NIH3T3 cells were transfected with plasmids encoding p110β and p85, or with plasmid encoding 12V-Ras. Cells were incubated with or without p110β-peptide or scrambled peptide, and formation of colonies in soft agar was measured. (F) Migration of NIH3T3 stably expressing p110β and p85α towards FBS in a Boyden chamber, in the absence or presence of p110 β-peptide or scrambled peptide. The graphs in each panel show means ± SEM from three to four separate experiments. Data in (E) show means ± SEM from triplicate measurements and are representative of two experiments.
Control experiments showed that the effects of the myristoylated peptide were specific for p110β-Gβγ interactions. The myristoylated peptide did not reduce the abundance of p110β protein (Fig. 5, C and D). In addition, the myristoylated p110β peptide had no effect on Gβγ-dependent activation of the class IB PI3K (the p101–p110γ dimer) (fig. S8A), the synergistic activation of adenylyl cyclase by Gβγ and Gαs (fig. S8B), or the Gβγ-mediated activation of PLC-β in cells (fig. S8, C), and the non-modified peptide had no effect on Gβγ-mediated activation of PLC-β in vitro (fig. S8, D). Similarly, the myristoylated peptide had no effect on the binding of p110β to Rab5 or on the p110β-dependent induction of autophagy (fig. S8, E and F) [29], which is in agreement with the 532KK-DD mutant p110β having no effect on Rab5 binding (Fig. 2C). Thus, the effects of the myristoylated peptide specifically disrupted p110β-Gβγ interactions. These data showed that p110β-mediated cellular transformation and migration requires the binding of p110β to G βγ.
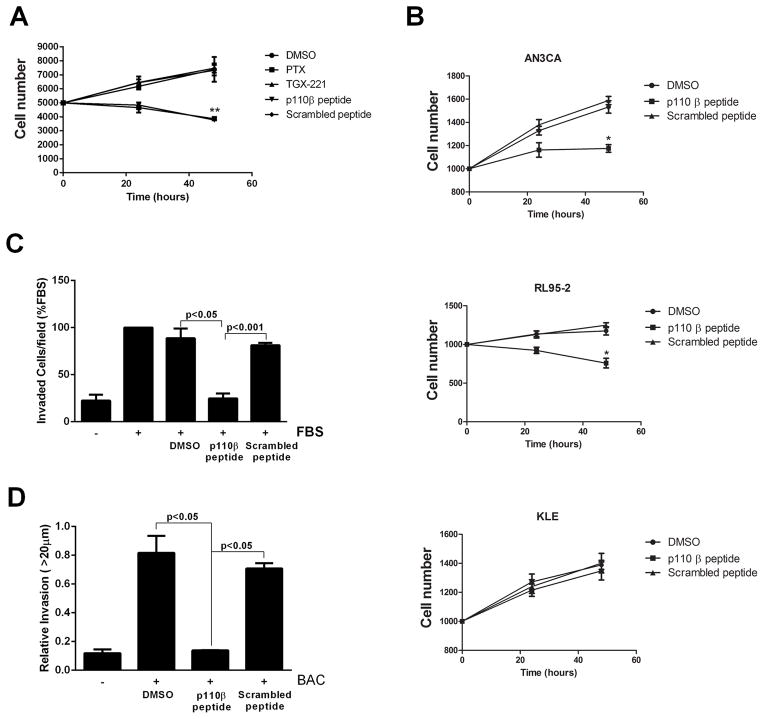
The growth of PTEN-null tumors depends on p110β [8], and inhibition of Gβγ signaling or knock-in of a kinase-deficient p110β blocks the growth of prostate cancer cells [9, 30]. To test the role of p110β-Gβγ interactions in PTEN-null prostate cancer cells, we measured the proliferation of PC3 cells in the presence of myristoylated p110β-peptide or scrambled peptide. Whereas PC3 cell proliferation was unaffected by the myristoylated scrambled peptide or by the p110β inhibitor TGX221, proliferation was inhibited in the presence of myristoylated p110β-peptide or pertussis toxin (Fig. 6A). Similar effects were seen in the PTEN-null endometrial cancer cell lines AN3CA and RL95-2, but not in the PTEN-replete endometrial cancer line KLE (Fig. 6B). Myristoylated p110β-peptide also inhibited the chemotaxis of PC3 cells toward serum in a Boyden chamber assay (Fig. 6C). Finally, in a collagen invasion assay designed to mimic paracrine interactions between macrophages and tumor cells during invasion [31], macrophage-dependent PC3 cell invasion was blocked by the myristoylated p110β-peptide (Fig. 6D), but not by myristoylated scrambled peptide. These data suggest that GPCR-mediated activation of p110β in PTEN-null cells plays a critical role in proliferation, chemotaxis, and in paracrine interactions between tumor cells and macrophages during invasion.
Fig. 6.
Inhibition of the proliferation and chemotaxis of prostate cancer cells. (A) Proliferation of PC-3 cells was measured by the MTT assay in the absence or presence of 200 nM TGX221, 30 μM myristoylated p110β-derived peptide, or 30 μM scrambled peptide. (B) Proliferation assays were performed on two PTEN-null endometrial cancer cell lines (AN3CA and RL95-2 cells) and one PTEN-positive endometrial cancer cell line (KLE cells) grown in the absence or presence of myristoylated p110β-derived peptide or scrambled peptide. (C) Chemotaxis of PC-3 cells towards 10% FBS in the absence or presence of 20 μM p110β-derived peptide or scrambled peptide was measured in Boyden chambers. (D) Bone marrow–derived macrophages and Cell-tracker-red–labeled PC-3 tumor cells were co-plated in 24-well dishes and overlaid with collagen. Cells were incubated for 24 hours in the absence or presence of p110β-derived peptide or scrambled peptide, and invasion into the collagen was measured by confocal microscopy. The data are means ± SD from two separate experiments for (B) and (D), and are means ± SEM from three separate experiments for (A) and (C).
Discussion
Over the last few years, there has been an increased appreciation of the roles of GPCRs in cancer, both through direct signaling and by transactivation of RTKs [32–34]. Although the activation of the PI3Kβ isoform of PI3K by Gβγ subunits has been known for many years, the mechanism of this interaction is unclear, and it has been difficult to specifically study GPCR-regulated signaling by PI3Kβ. Our identification of the Gβγ-binding site in p110β, and the reciprocal p110β-binding site in Gβγ, has enabled the construction of mutants and peptide-based inhibitors that specifically disrupt this interaction. Using these approaches, we have demonstrated a critical role for Gβγ signaling to PI3Kβ in p110β-mediated transformation, as well asin the proliferation and invasion of PTEN -null prostate cancer cells. Our data suggest that GPCR-mediated activation of PI3Kβ could provide a new target for the design of anticancer therapeutics.
The Gβγ-binding site comprises a surface loop that bridges helices Lα5 and H1A between the C2 and helical domains of p110β. This loop is close to the inhibitory contact site for the nSH2 domain of p85 (Glu552). We can propose two mechanisms for the activation of p110β by Gβγ: one through membrane recruitment, and the other through relief of SH2-mediated inhibition. These mechanisms are not mutually exclusive, and it is likely that both contribute. Gβγ stimulates p110β in the absence of p85 [7], as well as when p110β was associated with a p85 construct consisting of only the iSH2 domain (p85-i) or with a construct possessing the iSH2 connected to the inhibitory cSH2 domain (p85 -ic) (fig. S9). Furthermore, Gβγ activated PI3Kβ in the absence of pY (Fig. 1F). Consequently, relief of SH2-mediated inhibition cannot be the only mechanism of activation of p110β by Gβγ. Our studies with PI3Kβ and Gβγ in the presence of liposomes showed that Gβγ-binding enhanced interactions of the kinase domain with lipid membranes (fig. S2, B and D). This suggests that a portion of the activation mechanism involves increased targeting to membranes because of the lipid moiety of the prenylated Gβγ. On the other hand, activation of p110β by pY peptides involves the relief of inhibition by the N- and C-terminal SH2 domains [15], and both pY and Gβγ are required for maximal stimulation of PI3Kβ. It is possible that pY binding to the nSH2 only partially relieves its inhibitory contact, and that Gβγ more completely displaces it to achieve full activation. It is also possible that by Gβγ increasing the membrane affinity, the presence of the membrane surface sterically helps to displace the inhibitory N- and C-terminal SH2 domains. Of note, the Gβγ-binding region of p110β shows low sequence similarity with the corresponding region of the other Gβγ-regulated PI3K catalytic subunit, p110γ. Consequently, it is not straightforward to predict the Gβγ-binding region of p110γ, and this will require experimental mapping.
Because the peptide inhibitor did not affect the activity of p110β directly, we presume that its mechanism of action is through binding to the p110β-interacting site within Gβγ. HDX-MS analysis of p110β binding to Gβγ revealed a partial overlap with surfaces that bind to canonical Gβγ effectors, as well as a region that appears to be unique to p110β: the linker region between the N-terminal α-helix and the first blade of Gβ. We have not determined the binding site for the p110β-derived peptide within Gβγ. However, the specificity of the peptide’s effects for the interaction between Gβγ and p110β, rather than to adenylyl cyclase, PLC-β or p101–p110γ, suggests that the peptide interacts with the unique region of the p110β-binding surface in Gβγ. An alternative explanation accommodates the fact that the binding of p110β to Gβγ is weak relative to that of other canonical effectors. In this model, the p110β peptide may bind to a portion of the canonical interface, but with an affinity low enough to displace p110β, but not other Gβγ effectors. We cannot experimentally distinguish between these hypotheses at this time. Finally, it is formally possible that the peptide contacts Gβγ in a manner that is distinct from that which occurs with the corresponding loop in p110β. Studies are in progress to define the peptide-binding site, and these will be useful in designing a better inhibitor of the Gβγ-p110β interaction.
PI3Kβ is ubiquitously-expressed and has been implicated in the regulation of vascular tone [35], thrombogenesis [36], male fertility [37], phagocytosis in macrophages [38], and integrin signaling [39]. In addition, p110β has kinase-independent functions, including involvement in clathrin-mediated endocytosis, cell proliferation, and DNA repair [10, 40, 41]. The role of GPCR signaling to PI3Kβ in these systems can now be directly addressed. With regard to the requirement for PI3Kβ in PTEN-null tumors [8], our data suggest that Gβγ interactions with PI3Kβ are critical for the growth and invasion of these tumors. Surprisingly, the peptide was more efficacious in inhibiting the proliferation of PC3 cells than was the p110β-specific kinase inhibitor TGX221. This is consistent with studies showing that kinase-deficient p110β rescues proliferative defects in mice [10, 40], and suggests that at least some of the Gβγ signaling to p110β involves the scaffolding functions of p110β. In contrast, previous studies have shown that kinase-deficient p110β does not support transformation in PTEN-null cells [8], suggesting that stimulation of PI3Kβ activity by Gβγ is required for transformation.
The role of GPCR signaling in PTEN-null tumors has not been extensively studied. It will be important to determine whether peptidomimetics or other small-molecule inhibitors of the p110β-Gβγ interface might be therapeutically useful in the treatment of some PTEN-null tumors. Currently, we do not know which GPCRs function upstream of Gβγ in the activation, targeting, or both of PI3Kβ. Defining these upstream inputs would provide an alternative approach to the treatment of tumors dependent on p110β.
Materials and Methods
Design and cloning of constructs and transfections
The 532KK-DD mutant was generated with the Quick-change kit (Stratagene). The Gβ-p85α-icSH2 fusion construct was cloned with standard digestion and ligation strategies, linking the sequence encoding the C-terminus of human Gβ1 to that encoding the N-terminus of human p85α-ic (residues 432 to 724) with a 25-residue linker of the following sequence: GSPGISGGGGGPGSGGGGSGGGGSG. All mutants were confirmed by sequencing. Transfections were performed with Fugene HD (Roche).
Purification of p110β-p85α dimers expressed in insect cells
Recombinant baculoviruses were generated and propagated with the Bac-to-Bac expression system (Invitrogen) according to the manufacturer’s recommendations. For expression, 3 L of Spodoptera frugiperda (Sf9) cells at a density of 1.0 × 106 cells/ml were co-infected with an optimized ratio of viruses encoding complexes of the catalytic and regulatory subunit of PI3K. After 55 hours of infection at 27°C, cells were harvested and washed with ice-cold phosphate-buffered saline (PBS) supplemented with 0.5 mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF; (Melford). Subsequently, cells were lysed by sonication for 4 min in 120 ml of buffer A1 [20 mM Tris (pH 8), 300 mM NaCl, 10 mM imidazole] containing 0.5 mM AEBSF, and were centrifuged for 20 min at 140,000_g_. The supernatant was filtered through a 0.45-μm Minisart filter unit (Sartorius Biotech) before loading onto two connected 5-ml HisTrap FF columns (GE Healthcare). The columns were washed first with buffer A1 and then with buffer A2 [20 mM Tris (pH 8), 100 mM NaCl, 10 mM imidazole, 2 mM 2-mercaptoethanol (2-ME)], and eluted with a gradient from 0 to 100% of buffer A2 containing 150 mM imidazole. Fractions were analyzed on 4 to 12% Bis-Tris Novex gels (Invitrogen) with MOPS buffer. The protein complex was further purified on a 5-ml HiTrap Q-HP column (GE Healthcare) with buffer C [20 mM Tris (pH 8), 2 mM dithiothreitol (DTT)], and was eluted with buffer D [20mM Tris (pH 8), 2 mM DTT, 1 M NaCl]. The complex was concentrated with AMICON 50K centrifugal filters (Millipore) and loaded onto a 16/60 Superdex 200 gel filtration column (GE Healthcare) at 4°C running with buffer E [20 mM Hepes (pH 7.5), 100 mM NaCl, 2 mM Tris(2-carboxyethyl)phosphine (TCEP)]. The heterodimer was concentrated to about 5 mg/ml, frozen in liquid nitrogen, and stored at −80°C.
Purification of p110β-p85α dimers from mammalian cells
HEK 293T cells were cotransfected with plasmids encoding myc-p110β and p85α, and the proteins were co-immunoprecipitated with anti-myc antibody. Pellets were washed sequentially three times in PBS containing 1% NP-40, three times in 50 mM Tris(pH 7.4), 500 mM LiCl2, and twice in 20 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA. Pellets were resuspended in a final volume of 50 μl of 40 mM HEPES (pH 7.4), 0.1 % bovine serum albumin, 1 mM EGTA, 7 mM MgCl2, 120 mM NaCl, 1 mM DTT, and 1 mM β-glycerophosphate.
Purification of Gβγ expressed in insect cells
Recombinant human Gβ1, N-terminally hexahistidine-tagged bovine wild-type Gγ2, and the Gγ2(C68S) mutant were produced in Sf9 cells and purified as described previously [28]. Isoprenylated Gβ1His-γ2 was isolated from the membrane fraction. The membrane extract was clarified by ultracentrifugation at 100,000_g_ for 1 hour, and diluted five times with a buffer containing 20 mM Hepes-NaOH (pH 7.7), 100 mM NaCl, 0.1 % polyoxyethylene-10-lauryl ether (C12E10), and 10 mM 2-ME. The extract was supplemented with 25 mM imidazole and incubated with Ni2+-NTA Superflow beads (Qiagen) for 1 hour. The mixture was loaded onto a column cartridge and extensively washed with buffer containing 20 mM imidazole. Thereafter, bound insect Gα subunits were eluted withAlCl3in the presence of Mg 2+. Subsequently, Gβ1His-γ2 dimers were eluted with a buffer containing 20 mM Tris-HCl (pH 8.0), 25 mM NaCl, 0.1 % C12E10, 200 mM imidazole, and 10 mM 2-ME. Gβ1His-γ2 eluted from the Ni2+-NTA matrix was diluted and loaded onto a 1-ml Resource 15Q HR 5/5 column (GE Healthcare) equilibrated with a buffer containing 20 mM Tris-HCl (pH 8.0), 8 mM CHAPS, and 2 mM DTT. Bound proteins were eluted and fractionated with a continuous gradient elution (0 to 500 mM NaCl). Peak fractions were pooled and concentrated with Amicon 10 concentrators (Millipore). The protein was then loaded onto a gel filtration Superdex 200 HR 10/30 column (GE Healthcare) and eluted with a buffer containing 20 mM Hepes-NaOH (pH 7.7), 100 mM NaCl, 10 mM CHAPS, and 2 mM TCEP. Peak fractions were pooled and concentrated with Amicon 10 concentrators (Millipore). Purified proteins were quantified by Coomassie Brilliant Blue staining after SDS-PAGE analysis with BSA as a standard. Proteins were stored at −80°C. Non-lipidated Gβ1His-γ2(C68S) was purified from the cytosolic fraction of Sf9 cells. After separation from the membrane fraction, the cytosolic fraction was supplemented with 15 mM imidazole and incubated with Ni2+-NTA Superflow beads (Qiagen) for 1 hour. The mixture was loaded onto a column cartridge and extensively washed with a buffer containing 20 mM Hepes-NaOH (pH 7.7), 300 mM NaCl, 15 mM imidazole, and 10 mM 2-ME. Gβ1His-γ2(C68S) mutants were eluted with a buffer containing 20 mM Tris-HCl (pH 8.0), 25 mM NaCl, 200 mM imidazole, and 10 mM 2-ME. The protein eluted from the Ni2+-NTA matrix was diluted and loaded onto a 1-ml Resource 15Q HR 5/5 column (GE Healthcare) equilibrated with a buffer containing 20 mM Tris-HCl (pH 8.0) and 2 mM DTT. Bound proteins were eluted and fractionated with a continuous NaCl gradient elution (0 to 600 mM NaCl). Peak fractions were pooled and concentrated with Amicon 10 concentrators (Millipore). The protein was then loaded onto a gel filtration Superdex 200 HR 10/30 column (GE Healthcare) and eluted with a buffer containing 20 mM Hepes-NaOH (pH 7.7), 100 mM NaCl, and 2 mM TCEP. Peak fractions were pooled and concentrated with Amicon 10 concentrators (Millipore). Purified proteins were quantified by Coomassie Brilliant Blue staining following SDS-PAGE with BSA as a standard. Proteins were stored at −80°C.
Preparation of lipid vesicles
For assays with immunopurified material from mammalian cells, lipid vesicles consisting of 38% phosphatidylethanolamine (PE), 35.5% phosphatidylserine (PS), 16.3% phosphatidylcholine (PC), 3.5% sphingomyelin, and 6.7% phosphatidylinositol-4,5-bisphosphate (PIP2) (all percentages by weight) [42] were dried under argon, resuspended at 0.66 μg/μl in 40 mM HEPES (pH 7.4), 0.1 % BSA, 1 mM EGTA, 7 mM MgCl2, 120 mM NaCl, 1 mM DTT, and 1 mM β-glycerophosphate and were sonicated in a Branson cup sonicator. For assays with recombinant protein purified from insect cells, vesicles were prepared by adding the lipid components together in chloroform and evaporating the organic solvent under a stream of dry argon. The lipid film was allowed to dry for 30 min under vacuum, and was then resuspended in a solution of 20 mM Tris (pH 7.5), 100 mM KCl, 1 mM EGTA. The lipids were first bath-sonicated for 10 min, and then subjected to 10 cycles of freeze-thaw between liquid nitrogen and a 37°C water bath. The liposomes were finally extruded ten times through a 100-nm filter (Whatman, Anotop 10) with a gas-tight syringe. Vesicles were frozen at −80°C for storage and were used within 1 month of preparation. Vesicle composition was 5% Brain-PIP2 (Sigma), 20% Brain-PS (Sigma), 45% brain-PE (Avanti), 15% Dioleoyl-PC (Avanti), 10% cholesterol (Sigma), 5% egg-sphingomyelin (Sigma). Percentages are based on weight.
Assay of lipid kinase activity with immunopurified enzyme
For assays with immunoprecipitated enzymes, myc-tagged wild-type or 532KK-DD mutant p110β together with p85α were coimmunoprecipitated with an anti-myc antibody from appropriately transfected HEK 293T cells. For assays with Gβγ, Gβγ was preincubated with lipid vesicles for 30 min and then added to the resuspended enzyme pellets [44]. For assays with phosphopeptide, 1 μM (final concentration) tyrosyl phosphorylated peptide [mouse PDGFR residues 735 to 767, sequence: ESDGG(pY)MDMSKDESID(pY)VPMLDMKGDIKYADIE; referred to as pY] and lipid vesicles were added directly. The assay (immunoprecipitated enzyme and 200 nM Gβγ, 320 μM PE, 300 μM PS, 140 μM PC, 30 μM Sphingomyelin, and 300 μM PI in a final volume of 81 μl) was initiated by the addition of 5 μl of ATP (116 μM final concentration) containing 1 μCi [32P]ATP. After 10 min at 22°C, the assay was stopped by the addition of EDTA (50 μM final concentration), and 5-μl aliquots were spotted on nitrocellulose membranes. The membranes were washed 5 times in 1 M NaCl, containing 1% phosphoric acid, dried, and counted with a Molecular Dynamics Phosphorimager. Alternatively, assays were analyzed by thin layer chromatography by stopping the reaction with 20 μl of 7N HCl, mixing with 160 μl of a 1:1 solution of methanol:chloroform, and centrifugation to separate the phases, after which, 20μl of the organic phase was spotted onto a silica gel plate (EMD Merck). Plates were developed in a solvent system consisting of 60 ml of chloroform, 47 ml of methanol, 11.2 ml water, and 2 ml of ammonium hydroxide, dried, and counted with a Molecular Dynamics Phosphorimager. For assays using the inhibitory peptides, a 1 μM final concentration of peptides (WT p110β: KAAEIASSDSANVSSRGGKKFLPV; scrambled p110β: NGAEKVGSADSKSIAFVSLKARSP) in 20 mM Tris-HCl (pH 7.4), 10 mM NaCl was incubated with 200 nM of Gβγ for 30 min on ice, and then with lipids, as described earlier, for 10 min on ice, and finally for 10 min with immunopurified PI3K, after which the kinase assay described earlier was performed. For assays of immunopurified PI3Kγ in the presence of peptide, peptides (1 μM WT p110β; 1 μM scrambled p110β; 10 μM SIGK: SIGKAFKILGYPDYD; or 1 μM QEHA: QEHAQEPERQYMHIGTMVEFAYALVGK) in 20 mM Tris-HCl (pH 7.4), 10 mM NaCl was incubated with 200 nM of Gβγ for 30 min on ice, and was then incubated with lipids as described earlier for 10 min on ice, before finally being incubated for 10 min with immunopurified PI3Kγ from baculovirus-infected insect cells, after which the kinase assay described above was performed.
Assay of lipid kinase activity with enzyme purified from insect cells
For assays with recombinant PI3K from baculovirus-infected insect cells, lipid vesicles were used at a final concentration of 1 mg/ml and were prepared as described earlier. Stock solutions of three-fold concentrated WT or mutant PI3Kβ constructs were prepared at 75 nM (for assays of basal, pY-stimulated, and Gβγ-stimulated activity) and at 0.75 nM (for assay of synergistic activation by pY and Gβγ) in 20 mM Hepes (pH 7.5), 100 mM NaCl, 2 mM DTT, 9 mM MgCl2, 3 mM EDTA. Substrate stock solutions containing lipids (3 mg/ml) supplemented with either 900 nM RTK-pY [from a 100 μM stock in 10 mM Hepes (pH 7.5), 0.2% DMSO], 1.5 μM Gβ1γ2 [from a 50 μM stock in 20 mM Hepes (pH 7.5), 100 mM NaCl, 2 mM TCEP, 10 mM CHAPS], or both agonists were prepared in 20 mM Hepes (pH 7.5), 100 mM NaCl, 2 mM DTT. The concentrations of CHAPS and DMSO were adjusted to be equal under all conditions. A 300 μM ATP solution containing 0.1 mCi/ml of [γ32P]-ATP was prepared. The reaction was started by mixing 3 μl of protein stock with 3 μl of substrate stock and 3 μl of ATP solution. The reaction was stopped after 60 min by transferring 3 μl of reaction mixture to 3 μl of a 20 mM EDTA quench buffer. Lipid kinase activity was determined with a modified membrane-capture radioactive assay measuring the production of 32P-labeled PIP3 [43]. Three microliters of this mixture was then spotted on a nitrocellulose membrane. The membrane was dried and washed six times with 1 M NaCl, containing 1% phosphoric acid. The membrane was then air-dried before exposure to a phosphor screen (Molecular Dynamics) for 15 min. The intensity of the spots on the membrane was imaged with a Typhoon Phosphorimager (GE Healthcare) and quantified with ImageQuant software (GE Healthcare).
HDX-MS measurements
HDX-MS analyses of PI3Kβ and Gβγ were performed by following a similar protocol as that previously described [16]. In the experiment identifying interaction sites between PI3Kβ and soluble Gβγ-C68S, the rate of exchange of the p110β–Gβ1-p85α-icSH2–Gγ2-C68S fusion heterotrimer was compared to those of a p110β–p85α-icSH2 free heterodimer and a free Gβ1γ2-C68S heterodimer. Protein stock solutions at 7 μM were prepared in 20 mM Hepes (pH 7.5), 100 mM NaCl and 2 mM DTT. Exchange reactions were started by mixing 10 μl of protein stock with 40 μl of a 98% D2O solution containing 10 mM HEPES (pH 7.5), 50 mM NaCl, reaching a final concentration of 78% D2O. Deuterium exchange reactions were run for 3, 30, 300, and 3000 s of on-exchange at 23°C before the reactions were quenched. An additional experiment for 3 s of on-exchange was performed at 0°C to examine exchange rates of very rapidly exchanging hydrogens. On-exchange was stopped with 20 μl of quench buffer containing 1.2% formic acid and 2 M Guanidine-HCl, which lowered the pH to 2.6. Samples were then immediately frozen in liquid nitrogen and stored at −80°C for no longer than 7 days. For HDX-MS studies in the presence of lipids, on-exchange experiments were performed in the presence of 10 μM PDGFR pY. Lipid vesicles at 5 mg/ml were diluted 8-fold with the 98% D2O solution described earlier. Protein stock solutions containing 10 μM of pY [40 μM stock in 10 mM HEPES (pH 7.2), 0.08 % DMSO] were prepared and incubated for 10 min before the addition of deuterated buffer. To shift the equilibrium toward the PI3Kβ-lipidated Gβγ complex and minimize the concentration of free p110β-p85 heterodimer, the Gβγ concentration used (10 μM) was in excess of the PI3Kβ concentration (3 μM). PI3Kβ-pY (state 1), in the presence of lipids (state 2), and in the presence of lipids and Gβγ (state 3) were used in this set of experiments to differentiate between changes in the exchange of PI3Kβ arising from membrane interaction and those from Gβγ interaction. Exchange reactions were started by the addition of 10 μl of protein stock to 40 μl of lipid-containing D2O solution, reaching a final concentration of 69% D2O. Deuterium exchange reactions ran for the same time points described for experiments with the fusion construct, but no measurements were performed at 0°C because of problems with lipid precipitation. Samples were stored at −80°C for a maximum of 1 week before deuterium incorporation was measured. Every time point and state was a unique experiment, and every HDX-MS experiment was repeated twice.
Measurement of deuterium incorporation
Samples were rapidly thawed on ice and injected onto a ultra-performance liquid chromatography (UPLC) system immersed in ice. The protein was run over an immobilized pepsin column (Applied Biosystems, Poroszyme, 2-3131-00) at 130 μl/min, and collected over a particle van-guard pre column (Waters) for 3 min. The trap was then eluted in line with an Acquity 1.7 μm-particle, 100 mm × 1 mm C18 UPLC column (Waters) with a 5 to 36% gradient of buffer A (0.1% formic acid) and buffer B (100% acetonitrile) over 20 min, and injected onto a LTQ Orbitrap XL (Thermo Scientific) to acquire mass spectra of peptides ranging from 350 to 1500 m/z.
Protein digestion and peptide identification
Mass analysis of the peptide centroids was performed as described previously, using the software HD-Examiner (Sierra Analytics) [16]. Initial peptide identification was done by running tandem MS/MS experiments using a 5 to 35% B gradient over 60 min with an LTQ Orbitrap XL (Thermo Scientific). Peptides were identified by Mascot search in Thermo Proteome Discoverer software v. 1.2 (Thermo Scientific) based on fragmentation and peptide mass. The MS tolerance was set at 3 ppm, with a MS/MS tolerance of 0.5 Daltons. All peptides with a Mascot score >15 were analyzed by the HD-Examiner software. Any ambiguous peptides were excluded from the analysis. The full list of peptides was then manually validated by searching a non-deuterated protein sample MS scan to test for correct m/z state, and to check for the presence of overlapping peptides. The HD-Examiner software was used to automate the initial analysis of deuterium incorporation, but every peptide listed in the manuscript was manually verified at every state and time to check for correct charge state, m/z range, presence of overlapping peptides, and proper retention time.
Mass analysis of peptide centroids
Selected peptides were manually examined for deuterium incorporation and accurate identification. Results are presented as relative extent of deuteration with no correction for back exchange, because no fully deuterated protein sample could be obtained. However, a correction was applied to compensate for differences in the amount of deuterium in the exchange buffer (78 or 69% in experiments with lipids). The real extent of deuteration was ~25 to 35% higher than what is shown, based on tests performed with fully deuterated standard peptides. The average error was ≤0.2 Daltons for corrected data of two replicates. The deuterium incorporation was also plotted versus the on-exchange time. The 3 s at 0°C time point was labeled as 0.3 sec. Because we performed the experiments with lipids at lower protein concentration to increase the lipid to protein ratio, some peptides analyzed for the fusion construct could no longer be analyzed.
Akt activation
HEK 293T or HEK 293E cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and transfected with plasmids encoding human p85α, wild-type (WT) or mutant human myc-p110β, myc-Akt with or without plasmids encoding FLAG-tagged Gβ1 (FLAG-G β1) and hemagglutinin (HA)-tagged Gγ2 (HA-Gγ2), as indicated. Cells were incubated overnight in serum-free medium. N-myristoylated peptides (WT: KAAEIASSDSANVSSRGGKKFLPV; scrambled: NGAEKVGSADSKSIAFVSLKARSP; 50 mM stock in DMSO; final concentration: 30 μM), wortmannin (100 nM), PIK-75 (10 nM), and TGX-221 (50 nM) were added to the medium for 30 min before lysis of cells. After incubation, cells were lysed and subjected to immunoprecipitation with anti-myc antibodies. Lysates and immunoprecipitates were analyzed by Western blotting for Akt and pT308-Akt with specific antibodies (Cell Signaling Technologies), and were analyzed by ECL (GE Healthcare) followed by densitometry, or with the LICOR Odyssey imaging system. Results are shown as the ratio of the abundance of pAkt to that of total Akt.
Transformation assays
NIH 3T3 cells grown in DMEM containing 10% normal calf serum (NCS) were transfected with plasmids encoding p110β and p85α constructs. Two days after transfection, cells (2,500 cells/well) were plated in 1 ml of 0.3% top agar over 1 ml of 0.6% bottom agar in a six-well dish. Cell colonies were counted 3 weeks later. In assays with the myristoylated peptides, peptides were diluted to a concentration of 30 μM in both the top and bottom gels as well as in the media.
Focus formation assays
NIH 3T3 cells were plated (at 2 × 105 cells/well) in 6-well dishes and were transfected with plasmids encoding myc-p110β and p85α constructs Cells were grown for two weeks, with media changed every two days. The cells were fixed and stained with crystal violet, and the numbers of foci per well were counted. In assays with the myristoylated peptides, peptides were diluted to a concentration of 30 μM in the media for the duration of the assay.
Rab5 pulldown assays
HEK 293T cells were transfected with Fugene HD with plasmids encoding WT or mutant myc-p110β and p85α. The cells were washed with cold PBS and lysed in 120 mM NaCl, 20 mM Tris (pH 7.5), 1 mM MgCl2, 1 mM CaCl2, 10% glycerol, 1% NP40, containing EDTA-free Protease inhibitor cocktail (Roche) and Phosphatase inhibitor cocktails 1 (EMD) and 2 (Sigma). Lysates were incubated with GTPγS-Rab5 or glutathione-_S_-transferase (GST) beads as described previously [45]and washed, and bound proteins were eluted and analyzed by Western blotting.
Boyden chamber assays
NIH3T3 cells, NIH3T3 cells stably expressing WT or mutant p110β, or PC-3 cells were plated at 5 × 104 cells on tissue culture inserts containing 8.0 μm pores. The inserts were incubated with serum-free media in the presence of DMSO or myristoylated peptides (30 μM) in the upper chamber and media containing DMSO or peptides with 10% FBS in the lower chamber. After 24 hours, the cells were fixed in 4% paraformaldehyde. The insert membranes were removed, stained and mounted on coverslips with Dapi Fluoromount (Southern Biotech). Images were collected at 10x magnification with a Nikon Diaphot inverted fluorescence microscope and a SPOT Idea digital camera, and were analyzed using ImageJ software.
MTT cell proliferation assays
The MTT assay (Invitrogen) was performed as described by the manufacturer. Briefly, 1 × 103 cells were plated in 96-well plates in the appropriate medium with or without DMSO or 30 μM myristoylated peptides. At various times, the cells were incubated with a 12 mM MTT solution in PBS for 4 hours at 37°C. An equal volume of 0.1 g/ml SDS solution in 0.01 M HCl was added, and absorbance was read at 570 nm with a Spectramax M5 plate reader (Molecular Devices). The number of cells was calculated from the ratio of OD:cell number from a known number of cells on day1.
Collagen invasion Assay
BAC-1.2F5 macrophages and PC-3 tumor cells were vitally-labeled with CellTracker Red CMPTX and CellTracker Green CMFDA, respectively, and co-cultured at a 2.5:1 ratio in a MatTek plate. After cell attachment, the cells were overlaid with a collagen I gel. Invasion into the 3D gel was quantified after 24 hours by laser scanning confocal microscopy detection of the fluorescent signal from the red and green CellTracker dyes as described previously [31].
Adenylyl cyclase assay
Sf9 cells were infected with baculovirus coding for recombinant adenylyl cyclase 2. Sf9 cell membranes containing adenylyl cyclase 2 were prepared as previously described [46]. Adenylyl cyclase activity was measured with the procedure described by Smigel [47]. All assays were performed for 10 min at 30°C in a final volume of 100 μl containing 5 μg of cyclase-containing Sf9 membrane protein, 20 nM each of recombinant Gαs and Gβγ, and 30 μM of myristoylated p110β peptide or a previously described inhibitory QEHA peptide (QEHAQEPERQYMHIGTMVEFAYALVGK) [48]. The data are mean ± the standard deviation (SD)from duplicate determinations, and are representative of two separate experiments.
LC3 punctae assays
HEK 293A cells stably expressing green fluorescent protein (GFP)-tagged LC3 were plated on poly-L-lysine–coated coverslips, treated with DMSO or myristoylated peptides (30 μM) for 30 min, and then incubated in PBS, 100 nM rapamycin and peptide for 2 hours at 37°C. Coverslips were fixed in 4% PFA for 10 min at room temperature, and then imaged with 60× 1.4 N.A. optics with a Nikon Eclipse E400 microscope. Images were collected with a Roper cooled CCD camera and analyzed using ImageJ software.
In vitro activation of PLCβ
L-α-phosphatidylethanolamine (Avanti Polar Lipids, bovine liver), L-α-phosphatidylinositol-4,5-bisphosphate (Avanti, porcine brain) and [3H] phosphatidylinositol-4,5-bisphosphate (NEN Radiochemicals) were combined in chloroform, dried under a stream of N2, and resuspended in 20 mM Hepes (pH 7.2) by sonication. Recombinant PLCβ3 (1 nM) was incubated with the indicated concentrations of p110β peptide, scrambled peptide, or SIGK peptide, with or without 200 nM Gαq and in the presence or absence of w60 nM Gβγ, for 10 min at 30°C in a final volume of 60 μl containing 20 mM Hepes (pH 7.2), 8.3 mM NaCl, BSA (0.167 mg/ml), 2 mM DTT, 70 mM KCl, 3 mM EGTA, 10 mM NaF, 20 μM AlCl3, 5 mM MgCl2, 33 μM PIP2, 333 μM PE, 10,000 to 15000 dpm [3H]PIP2and CaCl 2added to give a free concentration of 200 nM Ca2+. The assay was terminated by the addition of 200 μl of 10% TCA and 100 μl of BSA (10 mg/ml), followed by centrifugation for 10 min at 4500 g. The supernatant was quantified by liquid scintillation spectrometry.
Quantification of [3H]inositol phosphate accumulation in cells
COS-7 cells were transiently transfected with or without plasmids encoding Gβγ subunits with Fugene 6. The culture medium was changed approximately 48 hours after plating to inositol-free DMEM (MP Biomedical) containing 1 μCi/well [2-3H(N)]myo-inositol (American Radiolabeled Chemicals). Metabolic labeling proceeded for 18 hours, at which point 100 μl of myristoylated or TAT-labeled peptide (to a final concentration of 30 μM) was added. After 30 min, 50 mM LiCl in 20 mM HEPES (pH 7.2) was added for 1 hour at 37°C. Incubations were terminated by aspiration of media and the addition of ice-cold 50 mM formic acid, followed by neutralization with 150 mM NH4OH after cell lysis. [3H]Inositol phosphates were isolated and quantified by Dowex chromatography. Parallel dishes were lysed and assayed for Akt activation as described earlier.
Statistical analysis
Error bars are show the SEM for experiments performed three or more times, and the SD for experiments performed twice. Statistical analyses were performed by ANOVA.
Supplementary Material
Figure S1
Fig. S1. Sequence alignment of p110α, p110β, and p110δ.
Figure S2
Fig. S2. Mapping of the Gβγ-binding region on p110β at the membrane with HDX -MS.
Figure S3
Fig. S3. Mapping of the Gβγ-binding region in p110β with HDX -MS.
Figure S4
Fig. S4. In vitro stimulation of PI3Kβ mutants by G βγ.
Figure S5
Fig. S5. Activation of Akt in cells transfected with plasmids encoding PI3Kβ and Gβγ: inhibition by TGX221 and p110β-derived peptide.
Figure S6
Fig. S6. Pertussis toxin–sensitive effects of PI3Kβ.
Figure S7
Fig. S7. Peptides derived from p110β inhibit GPCR -mediated activation of p110β signaling in intact cells.
Figure S8
Fig. S8. Peptide inhibitors of Gβγ-mediated PI3Kβ activation are specific for p85 -p110β.
Figure S9
Fig. S9. In vitro activity of heterodimers of p110β associated with p85 truncation constructs.
Supp. Figure Legends
Table S1
Table S1. Summary of all p110β peptides analyzed by HDX -MS for the p110β-p85α-icSH2 dimer (WT) and for the p110β-Gβ-p85α-icSH2-Gγ-C68S (fusion) complexes.
Table S2
Table S2. Summary of all p85 regulatory subunit peptides analyzed by HDX-MS for the p110β-p85α-icSH2 dimer (WT) and for the p110β-Gβ-p85α-icSH2-Gγ-C68S (fusion) complexes.
Table S3
Table S3. Summary of all peptide exchangedata for the G β subunit for HDX -MS experiments with WT and fusion complexes. γ
Table S4
Table S4. Summary of all peptide exchange data for the G subunit for HDX -MS experiments with WT and fusion complexes.
Table S5
Table S5. Summary of all p110β peptides analyzed by HDX -MS for PI3Kβ-pY with and without liposomes and Gβγ complexes.
Table S6
Table S6. Summary of all p85 regulatory subunit peptides analyzed by HDX-MS for the PI3Kβ-pY with and without liposomes and Gβγ complexes.
Acknowledgments
We thank C. S. Rubin, S. C. Almo, J. B. Bonnano, and P. Seneviratne for valuable discussions; M. Levy for the PC3 cell line; S. Tooze for the GFP-LC3 stable cell line; and S. B. Horwitz for the panel of endometrial cancer cell lines. We also thank F. Begum and S.-Y. Peak-Chew for help with the HDX-MS setup, J. Morrow for assistance with HD-examiner software, and R. Riehle for technical assistance with the purification of proteins.
Funding: O.V. was supported by a Swiss National Science Foundation fellowship (grant No PA00P3_134202) and a European Commission fellowship (FP7-PEOPLE-2010-IEF, N°275880). J.E B. was supported by an EMBO long-term fellowship (ALTF268-2009) and the British Heart Foundation (PG11/109/29247). H.A D. and B.D.K. were supported by grants from the Janey Fund. R.S S. was supported by NIH 5T32 GM007491 and by a National Research Service Award, 1 F31 AG040932-01. T.K.H. was supported by NIH grant GM57391. This work was funded by NIH grants GM55692 (to J.M.B). and PO1 CA 100324 (to J.M.B. and A.R.B),. by the Medical Research Council (to R.L.W,. file reference U105184308) and by Deutsche Forschungsgemeinschaft (to B.N).
Footnotes
Author contributions: H.A.D. identified the potential Gβγ interaction site by sequence analysis, produced mutants, analyzed mutant construct activity, performed cell culture analysis of p110β mutants, conducted peptide inhibition experiments for signaling, Boyden chamber and proliferation assays, prepared figures, and contributed to writing the paper. O.V. identified the potential Gβγ interaction site by HDX-MS, produced and purified PI3K mutants for in vitro activity assays and HDX-MS, carried out in vitro kinase assays, analyzed mutant construct activity, carried out all of the HDX-MS experiments, prepared figures, and contributed to writing the paper. A.S. expressed and purified proteins, including all of the Gβγ that was used for the HDX-MS, and conducted assays on recombinant p85/p110β. J.E.B. contributed to HDX-MS experiments and data analysis, and helped revise the manuscript. R.S.S. analyzed p110β binding to Rab5. B.D.K. conducted Boyden chamber experiments. M.O.B. performed cell-based PLCβ experiments. G.L.W. performed in vitro PLCβ experiments. C.S. performed binding experiments with peptides and Gβγ subunits. C.Hsueh performed collagen invasion assays. O.P. contributed to cloning, expression, purification, and activity assays of p110β constructs, and helped revise the manuscript. C.Harteneck contributed to expression, purification, and activity assays of Gβγ and PI3 -kinases. P.R.S. supervised the synthesis of the PI3K inhibitors. T.K.H. supervised the PLCβ experiments and provided purified Gβγ. A.V.S. supervised binding experiments with peptides and Gβγ subunits. R.T. conducted the adenylyl cyclase assays. A.R.B. contributed to the design and analysis of chemotaxis assays. B.N. provided purified Gβγ for kinase assays and HDX-MS experiments, and contributed to the design and analysis of assays on peptide and Gβγ effects on p110β activity of various PI3Kβ constructs, and contributed to writing of the manuscript. R.L.W. contributed to the design and analysis of the HDX-MS experiments and the p110β mutagenesis experiments, and contributed to writing the manuscript. J.M.B. contributed to the design and analysis of the p110β mutagenesis and the p110β activity and signaling experiments, and contributed to writing the manuscript.
Competing interests: H.A.D., R.S.S., B.D.K., and J.M.B. have a patent pending on the development of therapeutics targeting the p110β-Gβγ interface. P.R.S. is a founder scientist of Pathway Therapeutics.
References and Notes
- 1.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: Stabilization and inhibition of the p110-alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc Natl Acad Sci U S A. 2007;104:7809–7814. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni S, Sitaru C, Jakus Z, Anderson KE, Damoulakis G, Davidson K, Hirose M, Juss J, Oxley D, Chessa TA, Ramadani F, Guillou H, Segonds-Pichon A, Fritsch A, Jarvis GE, Okkenhaug K, Ludwig R, Zillikens D, Mocsai A, Vanhaesebroeck B, Stephens LR, Hawkins PT. PI3Kbeta plays a critical role in neutrophil activation by immune complexes. Sci Signal. 2011;4:ra23. doi: 10.1126/scisignal.2001617. [DOI] [PubMed] [Google Scholar]
- 5.Kurosu H, Maehama T, Okada T, Yamamoto T, Hoshino S, Fukui Y, Ui M, Hazeki O, Katada T. Heterodimeric phosphoinositide 3-kinase consisting of p85 and p110β is synergistically activated by the βgamma subunits of G proteins and phosphotyrosyl peptide. J Biol Chem. 1997;272:24252–24256. doi: 10.1074/jbc.272.39.24252. [DOI] [PubMed] [Google Scholar]
- 6.Murga C, Fukuhara S, Gutkind JS. A novel role for phosphatidylinositol 3-kinase beta in signaling from G protein-coupled receptors to Akt. J Biol Chem. 2000;275:12069–12073. doi: 10.1074/jbc.275.16.12069. [DOI] [PubMed] [Google Scholar]
- 7.Maier U, Babich A, Nürnberg B. Roles of non-catalytic subunits in Gβgamma-induced activation of class I phosphoinositide 3-kinase isoforms β and gamma. J Biol Chem. 1999;274:29311–29317. doi: 10.1074/jbc.274.41.29311. [DOI] [PubMed] [Google Scholar]
- 8.Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, Stegmeier F, Yao YM, Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berenjeno IM, Guillermet-Guibert J, Pearce W, Gray A, Fleming S, Vanhaesebroeck B. Both p110alpha and p110beta isoforms of PI3K can modulate the impact of loss-of-function of the PTEN tumour suppressor. Biochem J. 2012;442:151–159. doi: 10.1042/BJ20111741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, Zhang J, Signoretti S, Loda M, Roberts TM, Zhao JJ. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni J, Liu Q, Xie S, Carlson C, Von T, Vogel K, Riddle S, Benes C, Eck M, Roberts T, Gray N, Zhao J. Functional Characterization of an Isoform-Selective Inhibitor of PI3K-p110beta as a Potential Anticancer Agent. Cancer discovery. 2012;2:425–433. doi: 10.1158/2159-8290.CD-12-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien R, Rugman P, Renzoni D, Layton M, Handa R, Hilyard K, Waterfield MD, Driscoll PC, Ladbury JE. Alternative modes of binding of proteins with tandem SH2 domains. Protein Sci. 2000;9:570–579. doi: 10.1110/ps.9.3.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dbouk HA, Pang H, Fiser A, Backer JM. A biochemical mechanism for the oncogenic potential of the p110beta catalytic subunit of phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2010;107:19897–19902. doi: 10.1073/pnas.1008739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, Hawkins PT, Stephens LR, Williams RL. Structure of Lipid Kinase p110beta/p85beta Elucidates an Unusual SH2-Domain-Mediated Inhibitory Mechanism. Mol Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke JE, Vadas O, Berndt A, Finegan T, Perisic O, Williams RL. Dynamics of the phosphoinositide 3-kinase p110delta interaction with p85alpha and membranes reveals aspects of regulation distinct from p110alpha. Structure. 2011;19:1127–1137. doi: 10.1016/j.str.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA) Proc Natl Acad Sci U S A. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, Hawkins PT, Stephens LR, Williams RL. Structure of lipid kinase p110beta/p85beta elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang CH, Kinzler KW, Vogelstein B, Amzel LM. A frequent kinase domain mutation that changes the interaction between PI3Kalpha and the membrane. Proc Natl Acad Sci U S A. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun M, Hart JR, Hillmann P, Gymnopoulos M, Vogt PK. Addition ofN -terminal peptide sequences activates the oncogenic and signaling potentials of the catalytic subunit p110alpha of phosphoinositide-3-kinase. Cell Cycle. 2011;10:3731–3739. doi: 10.4161/cc.10.21.17920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier U, Babich A, Macrez N, Leopoldt D, Gierschik P, Illenberger D, Nurnberg B. Gbeta 5gamma 2 is a highly selective activator of phospholipid-dependent enzymes. J Biol Chem. 2000;275:13746–13754. doi: 10.1074/jbc.275.18.13746. [DOI] [PubMed] [Google Scholar]
- 24.Buck E, Li J, Chen Y, Weng G, Scarlata S, Iyengar R. Resolution of a signal transfer region from a general binding domain in gbeta for stimulation of phospholipase C-beta2. Science. 1999;283:1332–1335. doi: 10.1126/science.283.5406.1332. [DOI] [PubMed] [Google Scholar]
- 25.Panchenko MP, Saxena K, Li Y, Charnecki S, Sternweis PM, Smith TF, Gilman AG, Kozasa T, Neer EJ. Sites important for PLCbeta2 activation by the G protein betagamma subunit map to the sides of the beta propeller structure. J Biol Chem. 1998;273:28298–28304. doi: 10.1074/jbc.273.43.28298. [DOI] [PubMed] [Google Scholar]
- 26.Scott JK, Huang SF, Gangadhar BP, Samoriski GM, Clapp P, Gross RA, Taussig R, Smrcka AV. Evidence that a protein-protein interaction ‘hot spot’ on heterotrimeric G protein betagamma subunits is used for recognition of a subclass of effectors. EMBO J. 2001;20:767–776. doi: 10.1093/emboj/20.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Sternweis PM, Charnecki S, Smith TF, Gilman AG, Neer EJ, Kozasa T. Sites for Galpha binding on the G protein beta subunit overlap with sites for regulation of phospholipase Cbeta and adenylyl cyclase. J Biol Chem. 1998;273:16265–16272. doi: 10.1074/jbc.273.26.16265. [DOI] [PubMed] [Google Scholar]
- 28.Shymanets A, Ahmadian MR, Kossmeier KT, Wetzker R, Harteneck C, Nurnberg B. The p101 subunit of PI3Kgamma restores activation by Gbeta mutants deficient in stimulating p110gamma. Biochem J. 2012;441:851–858. doi: 10.1042/BJ20111664. [DOI] [PubMed] [Google Scholar]
- 29.Dou Z, Chattopadhyay M, Pan JA, Guerriero JL, Jiang YP, Ballou LM, Yue Z, Lin RZ, Zong WX. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol. 2010;191:827–843. doi: 10.1083/jcb.201006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bookout AL, Finney AE, Guo R, Peppel K, Koch WJ, Daaka Y. Targeting Gbetagamma signaling to inhibit prostate tumor formation and growth. J Biol Chem. 2003;278:37569–37573. doi: 10.1074/jbc.M306276200. [DOI] [PubMed] [Google Scholar]
- 31.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 32.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 33.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 34.Daaka Y. G proteins in cancer: the prostate cancer paradigm. Sci STKE. 2004;2004:re2. doi: 10.1126/stke.2162004re2. [DOI] [PubMed] [Google Scholar]
- 35.Macrez N, Mironneau C, Carricaburu V, Quignard JF, Babich A, Czupalla C, Nurnberg B, Mironneau J. Phosphoinositide 3-kinase isoforms selectively couple receptors to vascular L-type Ca(2+) channels. Circ Res. 2001;89:692–699. doi: 10.1161/hh2001.097864. [DOI] [PubMed] [Google Scholar]
- 36.Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CE, Kenche V, Anderson KE, Dopheide SM, Yuan Y, Sturgeon SA, Prabaharan H, Thompson PE, Smith GD, Shepherd PR, Daniele N, Kulkarni S, Abbott B, Saylik D, Jones C, Lu L, Giuliano S, Hughan SC, Angus JA, Robertson AD, Salem HH. PI 3-kinase p110beta: a new target for antithrombotic therapy. Nat Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 37.Ciraolo E, Morello F, Hobbs RM, Wolf F, Marone R, Iezzi M, Lu X, Mengozzi G, Altruda F, Sorba G, Guan K, Pandolfi PP, Wymann MP, Hirsch E. Essential role of the p110beta subunit of phosphoinositide 3-OH kinase in male fertility. Mol Biol Cell. 2010;21:704–711. doi: 10.1091/mbc.E09-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leverrier Y, Okkenhaug K, Sawyer C, Bilancio A, Vanhaesebroeck B, Ridley AJ. Class I phosphoinositide 3-kinase p110beta is required for apoptotic cell and Fcgamma receptor-mediated phagocytosis by macrophages. J Biol Chem. 2003;278:38437–38442. doi: 10.1074/jbc.M306649200. [DOI] [PubMed] [Google Scholar]
- 39.Jackson SP, Schoenwaelder SM. PI 3-Kinase p110beta regulation of platelet integrin alpha(IIb)beta3. Curr Top Microbiol Immunol. 2010;346:203–224. doi: 10.1007/82_2010_61. [DOI] [PubMed] [Google Scholar]
- 40.Ciraolo E, Iezzi M, Marone R, Marengo S, Curcio C, Costa C, Azzolino O, Gonella C, Rubinetto C, Wu H, Dastru W, Martin EL, Silengo L, Altruda F, Turco E, Lanzetti L, Musiani P, Ruckle T, Rommel C, Backer JM, Forni G, Wymann MP, Hirsch E. Phosphoinositide 3-kinase p110beta activity: key role in metabolism and mammary gland cancer but not development. Sci Signal. 2008;1:ra3. doi: 10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar A, Fernandez-Capetillo O, Carrera AC. Nuclear phosphoinositide 3-kinase beta controls double-strand break DNA repair. Proc Natl Acad Sci U S A. 2010;107:7491–7496. doi: 10.1073/pnas.0914242107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leopoldt D, Hanck T, Exner T, Maier U, Wetzker R, Nurnberg B. Gbetagamma stimulates phosphoinositide 3-kinase-gamma by direct interaction with two domains of the catalytic p110 subunit. J Biol Chem. 1998;273:7024–7029. doi: 10.1074/jbc.273.12.7024. [DOI] [PubMed] [Google Scholar]
- 43.Knight ZA, Feldman ME, Balla A, Balla T, Shokat KM. A membrane capture assay for lipid kinase activity. Nat Protoc. 2007;2:2459–2466. doi: 10.1038/nprot.2007.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leopoldt D, Hanck T, Exner T, Maier U, Wetzker R, Nurnberg B. Gbetagamma stimulates phosphoinositide 3-kinase-gamma by direct interaction with two domains of the catalytic p110 subunit. J Biol Chem. 1998;273:7024–7029. doi: 10.1074/jbc.273.12.7024. [DOI] [PubMed] [Google Scholar]
- 45.Christoforidis S, Zerial M. Purification and identification of novel Rab effectors using affinity chromatography. Methods. 2000;20:403–410. doi: 10.1006/meth.2000.0953. [DOI] [PubMed] [Google Scholar]
- 46.Taussig R, Tang WJ, Gilman AG. Expression and purification of recombinant adenylyl cyclases in Sf9 cells. Methods Enzymol. 1994;238:95–108. doi: 10.1016/0076-6879(94)38009-0. [DOI] [PubMed] [Google Scholar]
- 47.Smigel MD. Purification of the catalyst of adenylate cyclase. J Biol Chem. 1986;261:1976–1982. [PubMed] [Google Scholar]
- 48.Chen J, DeVivo M, Dingus J, Harry A, Li J, Sui J, Carty DJ, Blank JL, Exton JH, Stoffel RH, et al. A region of adenylyl cyclase 2 critical for regulation by G protein beta gamma subunits. Science. 1995;268:1166–1169. doi: 10.1126/science.7761832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Fig. S1. Sequence alignment of p110α, p110β, and p110δ.
Figure S2
Fig. S2. Mapping of the Gβγ-binding region on p110β at the membrane with HDX -MS.
Figure S3
Fig. S3. Mapping of the Gβγ-binding region in p110β with HDX -MS.
Figure S4
Fig. S4. In vitro stimulation of PI3Kβ mutants by G βγ.
Figure S5
Fig. S5. Activation of Akt in cells transfected with plasmids encoding PI3Kβ and Gβγ: inhibition by TGX221 and p110β-derived peptide.
Figure S6
Fig. S6. Pertussis toxin–sensitive effects of PI3Kβ.
Figure S7
Fig. S7. Peptides derived from p110β inhibit GPCR -mediated activation of p110β signaling in intact cells.
Figure S8
Fig. S8. Peptide inhibitors of Gβγ-mediated PI3Kβ activation are specific for p85 -p110β.
Figure S9
Fig. S9. In vitro activity of heterodimers of p110β associated with p85 truncation constructs.
Supp. Figure Legends
Table S1
Table S1. Summary of all p110β peptides analyzed by HDX -MS for the p110β-p85α-icSH2 dimer (WT) and for the p110β-Gβ-p85α-icSH2-Gγ-C68S (fusion) complexes.
Table S2
Table S2. Summary of all p85 regulatory subunit peptides analyzed by HDX-MS for the p110β-p85α-icSH2 dimer (WT) and for the p110β-Gβ-p85α-icSH2-Gγ-C68S (fusion) complexes.
Table S3
Table S3. Summary of all peptide exchangedata for the G β subunit for HDX -MS experiments with WT and fusion complexes. γ
Table S4
Table S4. Summary of all peptide exchange data for the G subunit for HDX -MS experiments with WT and fusion complexes.
Table S5
Table S5. Summary of all p110β peptides analyzed by HDX -MS for PI3Kβ-pY with and without liposomes and Gβγ complexes.
Table S6
Table S6. Summary of all p85 regulatory subunit peptides analyzed by HDX-MS for the PI3Kβ-pY with and without liposomes and Gβγ complexes.