Mapping cellular hierarchy by single cell analysis of the cell surface repertoire (original) (raw)
. Author manuscript; available in PMC: 2014 Apr 3.
Published in final edited form as: Cell Stem Cell. 2013 Sep 12;13(4):10.1016/j.stem.2013.07.017. doi: 10.1016/j.stem.2013.07.017
SUMMARY
Stem cell differentiation pathways are most often studied at the population level, whereas critical decisions are executed at the level of single cells. We have established a highly multiplexed, quantitative PCR assay to profile in an unbiased manner a panel of all commonly used cell surface markers (280 genes) from individual cells. With this method we analyzed over 1500 single cells throughout the mouse hematopoietic system, and illustrate its utility for revealing important biological insights. The comprehensive single cell dataset permits mapping of the mouse hematopoietic stem cell (HSC) differentiation hierarchy by computational lineage progression analysis. Further profiling of 180 intracellular regulators enabled construction of a genetic network to assign the earliest differentiation event during hematopoietic lineage specification. Analysis of acute myeloid leukemia elicited by MLL-AF9 uncovered a distinct cellular hierarchy containing two independent self-renewing lineages with different clonal activities. The strategy has broad applicability in other cellular systems.
INTRODUCTION
Cellular differentiation is commonly depicted as a sequential binary commitment process through multiple intermediate states. Using combinations of markers, different types of stem and progenitor cells have been identified in various systems. Further enrichment and analysis of these populations has aided appreciation of stepwise lineage specification. However, the choice of a small number of markers for enrichment of cell populations often masks potential heterogeneity and may bias an understanding of the cellular hierarchy.
Extensive cellular and molecular studies have contributed to the characterization of vertebrate hematopoietic differentiation pathways (Orkin and Zon, 2008). The prospective identification of mouse hematopoietic stem and progenitor cells (Muller-Sieburg et al., 1986; Visser et al., 1984), and further separation of hematopoietic stem (HSC) cells from multipotent progenitors (MPP) (Kiel et al., 2005; Morrison et al., 1997; Morrison and Weissman, 1994; Osawa et al., 1996), suggested a cellular hierarchy whereby self-renewing HSCs produce transiently amplifying multipotent progenitors (MPP). Subsequent identification of common lymphoid (CLP) and myeloid (CMP) progenitors (Akashi et al., 2000; Kondo et al., 1997) led to the conventional model in which lineage specification first takes place as a lymphoid (CLP) versus myeloid (CMP) bifurcation event. Several findings, however, challenge this simple view. They describe heterogeneity of early progenitor populations, and posit that lymphomyeloid lineage commitment may occur upstream of the separation of CLP and CMP (Adolfsson et al., 2005; Arinobu et al., 2007; Pronk et al., 2007). Different marker panels and FACS purification schemes have prevented resolution of these alternative models.
Cells within leukemias are also believed to form a hierarchy, yet descriptions of leukemia stem cells (LSC) are often seemingly contradictory. Original support for the existence of LSCs rested on the observation that only a rare subset of human acute myeloid leukemia (AML) cells, characterized by a surface phenotype similar to that of hematopoietic stem/progenitor cells (HSPCs), was competent to reinitiate disease upon transplantation in immunodeficient mice (Bonnet and Dick, 1997). More recent findings derived from a mouse model of AML driven by MLL-AF9 suggest that LSCs display a GMP-like phenotype and stand at the top of the leukemia hierarchy (Krivtsov et al., 2006). Other reports argue that leukemia cells with immunophenotypes of lineage cells may perform as functional LSCs in mouse AML (Gibbs et al., 2012; Somervaille and Cleary, 2006), adding to the complexity of the leukemia hierarchy.
Single cell gene expression analysis offers potential to resolve these issues. Recently, several hallmark technical advances have been achieved. Single cell mRNA sequencing strategies enable whole transcriptome analysis from individual cells (Islam et al., 2012; Ramskold et al., 2012; Tang et al., 2010; Tang et al., 2009). Alternatively, single cell mass cytometry constitutes a powerful system for multiplexed gene expression analysis at the protein level (Bendall et al., 2011). When both sample size and assayed gene number are taken into consideration, high-throughput single cell qPCR represents a favorable option (Buganim et al., 2012; Dalerba et al., 2011; Guo et al., 2010; Moignard et al., 2013). The qPCR approach is highly sensitive in detecting quantitative differences at mRNA level (Guo et al., 2010).
Here we sought to improve the utility and value of current single cell qPCR technology by increasing its throughput so as to assess expression of nearly all commonly used cell surface markers. We illustrate how this enhanced approach provides biological insights into normal and leukemic hematopoiesis. The approach we describe should be applicable to other developmental systems and allow for cross tissue, cross experiment comparisons. The method allows dissection of heterogeneous populations and the identification of cellular states at single cell resolution.
RESULTS
Single cell gene expression analysis of the cell surface repertoire
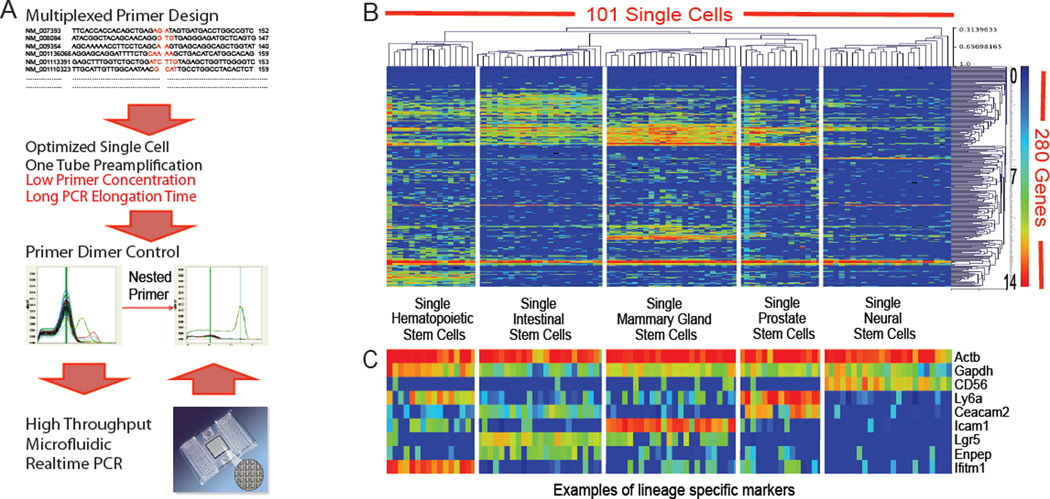
By introducing algorithm based primer design and optimizing the cycling conditions for highly multiplexed PCR, we have increased the capacity of single cell mRNA sequence specific pre-amplification (Figure 1A). In addition, the use of EvaGreen realtime PCR chemistry (Biotium) and melting curve analysis allows for non-specific signal control during gene specific qPCR on the BioMark realtime PCR system (Fluidigm). Finally, inclusion of nested primers filters out primer dimer signals (Figure 1A). In the highly multiplexed PCR pre-amplification, the chance of forming a dimer between a given primer pair of the same gene is actually very low. The subsequent gene specific qPCR will select and enrich target amplicons, even from extremely low starting materials (Figure S1A and S1B). We have designed and optimized a panel of assays to cover all commonly used cell surface markers (Lai et al., 1998) with a total of 280 genes (a few important transcription factors are also included) in establishing an analysis platform for all mouse cell types. After 280 multiplexed single cell pre-amplification, individual gene expression is quantified on the BioMark realtime PCR system (Fluidigm) using three 96.96 dynamic arrays.
Figure 1. Single cell gene expression analysis of the cell surface repertoire.
(A) Flow chart of single cell assay development. (B) A heatmap showing that the unbiased hierarchical clustering well separates single cell gene expression signatures from different type of adult stem cells. Each row corresponds to a specific gene; each column corresponds to a particular single cell. Red to yellow suggest high to middle expression, while green to blue suggest low to no expression. (C) A heatmap highlighting examples of lineage specific markers from Figure 1B. The color scale and sample layout are the same as in Figure 1B. See also Figure S1.
To assess the ability of the assay to discriminate different cell types at single cell level, we used flow cytometry to sort stem cell populations from a broad range of tissues including neural, prostate, mammary gland, intestinal and hematopoietic stem cells according to published protocols (Table S1A), and applied the single cell assay for gene expression profiling. As shown in Figure 1B, hierarchical clustering of the single cell data faithfully groups cells of the same origin together. The clustering also reveals lineage specific markers, such as CD56 for neural stem cells, Ceacam2 for prostate stem cells, Icam1 for mammary gland stem cells, Lgr5 for intestinal stem cells and Ifitm1 for hematopoietic stem cells (Figure 1C). False positive signal from a no cell pre-amplification control is extremely rare and weak (Figure S1C). These results provide initial evidence in behalf of the robustness of the single cell approach.
Comprehensive single cell analysis of the hematopoietic system
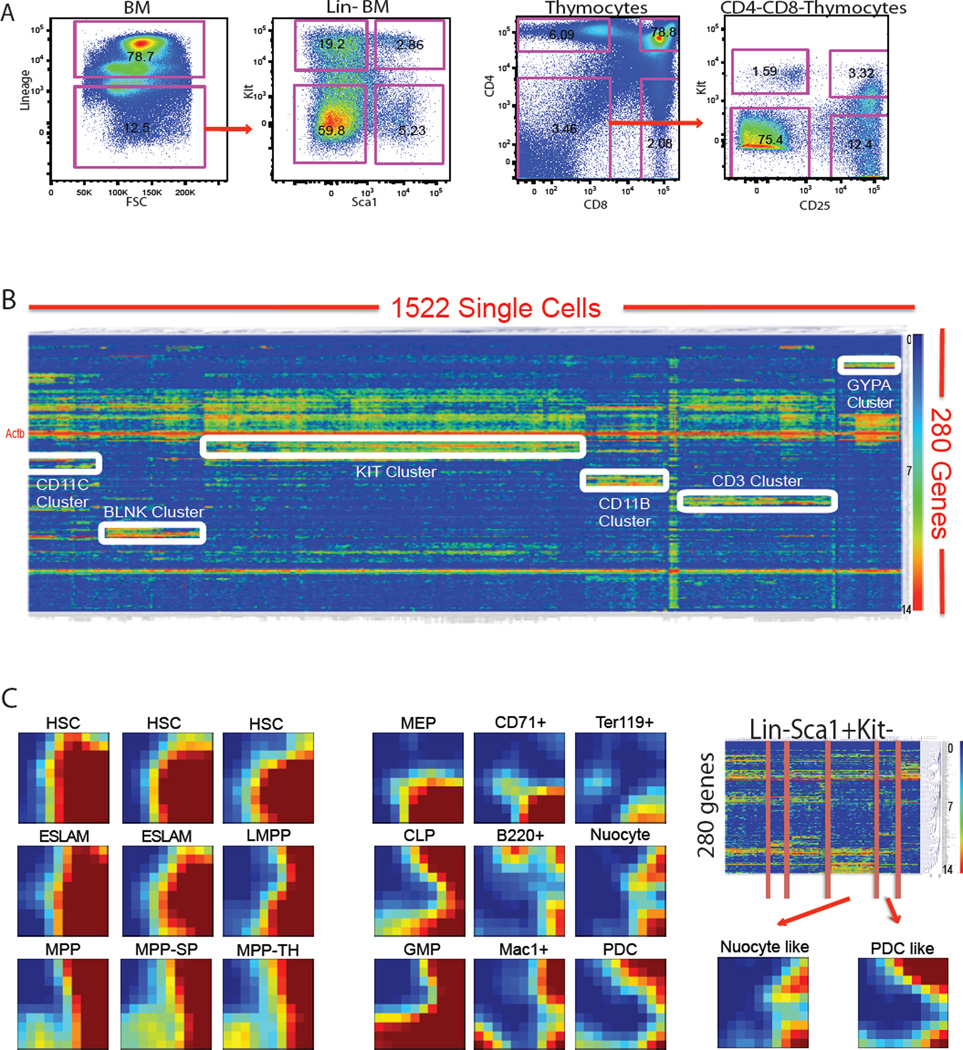
We utilized the single cell assay for a systematic analysis of the mouse hematopoietic system. To enrich stem cell and progenitor cell populations, and represent all possible cellular transitional states during differentiation, we used FACS to sort the principal hematopoietic compartments of the bone marrow by use of the cell surface markers c-Kit and Sca1, as well as a lineage (Lin) cocktail that recognizes mature cells of the major hematopoietic cell lineages, including T lymphocytes, B lymphocytes, monocytes/macrophages, granulocytes, and erythrocytes. Sorted populations include Lin+, Lin-Sca1+c-Kit+ (LSK), Lin-Sca1+c-Kit- (LSK-), Lin-Sca1-c-Kit+ (LS-K) and Lin-Sca1-c-Kit- (LS-K-) populations (Figure 2A). In addition, we sorted conventionally defined stem and progenitor cell types (including HSCs, MPP, CMP, CLP, common dendritic cell progenitor (CDP) (Onai et al., 2007), megakaryocyte/erythroid progenitor (MEP) and granulocyte/monocyte progenitor (GMP) as well as a set of differentiated cell types (Figure S2 and Table S1). A similar strategy was used to sort CD4+ T cells, CD8+ T cells, CD4+CD8+ double positive (DP) T cells, earliest thymic progenitors (ETP), CD4-CD8- double-negative (DN) 2, DN3 and DN4 stage thymocyte progenitors (Figure 2A, S2 and Table S1). An average of around 50 single cells are analyzed for each sorted population (Table S1). We analyzed more than 1500 single cells throughout the mouse hematopoietic system, and quantified all 280 genes for each individual cell (Table S2).
Figure 2. Comprehensive single cell analysis of the mouse hematopoietic system.
(A) Single cell sorting strategy to enrich stem and progenitor cells but to cover all possible populations. (B) A master heatmap showing the hierarchical clustering of gene expression signatures from 1500 single cells throughout the hematopoietic system. Each row corresponds to a specific gene; each column corresponds to a particular single cell. Strong correlation between gene and cell clusters are highlighted by white boxes and labeled by cell type specific clusters. Red to yellow suggest high to middle expression, while green to blue suggest low to no expression. (C) GEDI plot allows for visualization of single cell global signatures. Examples of single cell GEDI map from different cell types are presented. Color scale is as described in Figure 1B. The lower right corner, which is always red, corresponds to endogenous control genes that are highly expressed in all single cell samples. From the Lin-Sca1+Kit- population, there are cluster of single cells (The red lines separate different clusters in the heatmap of Lin-Sca1+Kit- single cell data) with Nuocyte signature and PDC signature. See also Figure S2.
Unsupervised hierarchical clustering of the single cell dataset reveals high correlation of gene expression clusters with cell type clusters (Figure 2B). As highlighted by white boxes, CD11c, CD3, Blnk, Kit CD11b and Gypa clusters correspond to dendritic, T, B, stem and progenitor, myeloid, megakaryocytic and erythroid (MegE) lineage cells, respectively. The principal lineage specific gene clusters are summarized in Table S3. Sub-clusters also exist within these main clusters. In addition, the quantified mRNA level differences correlate with different FACS sorting schemes; Actb and Gapdh expression levels are relatively consistent (Figure S3A). The clustering data suggest that differential global gene expression signatures at the single level are reproducible in both progenitor and differentiated cells types. The clustering pattern may then be used to identify novel markers and populations.
To visualize the overall pattern of gene expression (280 parameters) at the single cell level, we used the GEDI program (Chang et al., 2008) to generate individual expression maps (Figure 2C). The color of each pixel on the map indicates the centroid value of the gene expression level for each mini gene cluster generated by the software. Representative single cell maps from different populations illustrate how the method can be used to identify and classify virtually all cell types. As an example, we show that an incompletely defined Lin-Sca1+c-Kit- (LSK-) population is very heterogeneous as revealed by the clustering (Figure 2C). According to the single cell gene expression signature, this population contains not only CLP like progenitors and B cell progenitors, but also plasmacytoid dendritic cells (PDC) (Onai et al., 2007) and Nuocytes (Neill et al., 2010). Changes during cellular differentiation may be visualized from the maps. The gradual transition of the GEDI map from sorted MEP to CD71+ erythroid progenitors, and then to Ter119+ cells provides an example. Interestingly, we have identified bone marrow MPP like cells in the spleen (MPP-SP) and thymus (MPP-TH), consistent with the circulation of hematopoietic progenitor cells throughout the body.
Heterogeneity of hematopoietic progenitor cell types
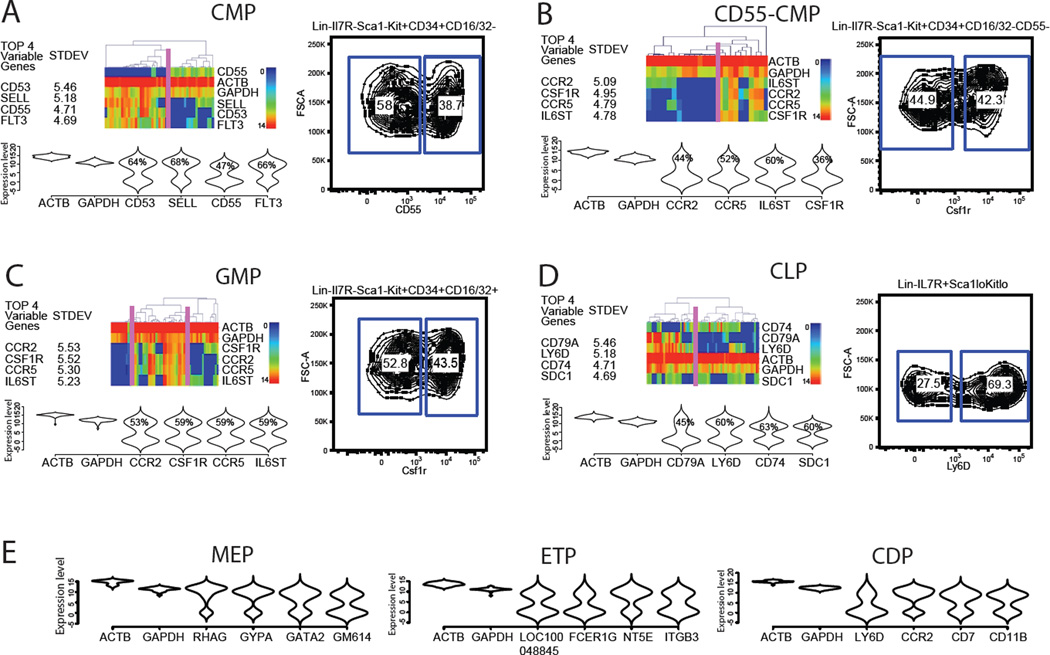
Having established a robust methodology for single cell analysis, we proceeded to examine the classically defined hematopoietic progenitor cell types (Figure 3A–3E). Each of these progenitor types reveals marked heterogeneity. For example, we profiled 47 single CMP cells, originally defined by the Lin-IL7R-Kit+Sca1-CD34+CD16/32lo profile, and ranked 280 genes by their standard deviation across all CMP samples. The top 4 most variable genes were CD53, Sell, CD55 and Flt3 (Figure 3A). Hierarchical clustering of these variable genes reveals two principal populations with different gene expression patterns. To address whether these gene expression differences reflect stochastic noise (Chang et al., 2008), we applied violin plot analysis to visualize the distribution of gene expression levels. In this plot, the Y- and X-axes correspond to the gene expression level and distribution frequency, respectively. Theoretically, expression noise should exhibit unimodal distribution around a reference level, whereas a multimodal distribution should indicate quantitative differences. As expected, the distributions of Actb and Gapdh levels are unimodal, with a very narrow peak indicative of low variation (Figure 3A and S3B). In contrast, the top 4 most variable genes within the CMP population show clear bimodal distribution. To confirm mRNA level differences at the protein level, we used available antibodies to the surface marker CD55 to further analyze the CMP compartment. Flow cytometry validated the heterogeneous nature of the CMP population detected by single cell qPCR (Figure 3A). We then continued to dissect heterogeneity further in the CD55-CMP population, and revealed Csf1r as one of most differentially expressed markers (Figure 3B). Comparable analyses were performed for GMP, CLP, MEP, ETP and CDP. We observed discrete heterogeneity within all populations (Figure 3C–3E). The analysis also reveals dynamic changes in LSK heterogeneity during the aging process (Figure S3C), and permits assessment of the purity of HSCs from different enrichment protocols (Figure S3D). The bimodal distribution of mRNA transcripts is present in all the cell types that we have purified, suggesting extensive unknown heterogeneities. Although the mRNA level expression is not always reflective of protein level expression, we argue that it should be indicative of a cell’s transcriptional state and functional potential.
Figure 3. Dissection of heterogeneity within classical progenitor types.
(A)–(D) Top 4 most variable genes are listed according to their standard deviation value within a particular progenitor cell type. The hierarchical clustering heatmap and violin density plot reveal the heterogeneity in the population. The percentages of cells with positive expression levels are marked on the violin plot. Color scale is as described in Figure 1B. FACS analysis confirms gene expression differences at protein level. (E) Violin plots showing the expression pattern of top 4 most variable genes in MEP, ETP and CDP progenitor populations. See also Figure S3.
Mapping hematopoietic hierarchy by computational lineage progression analysis
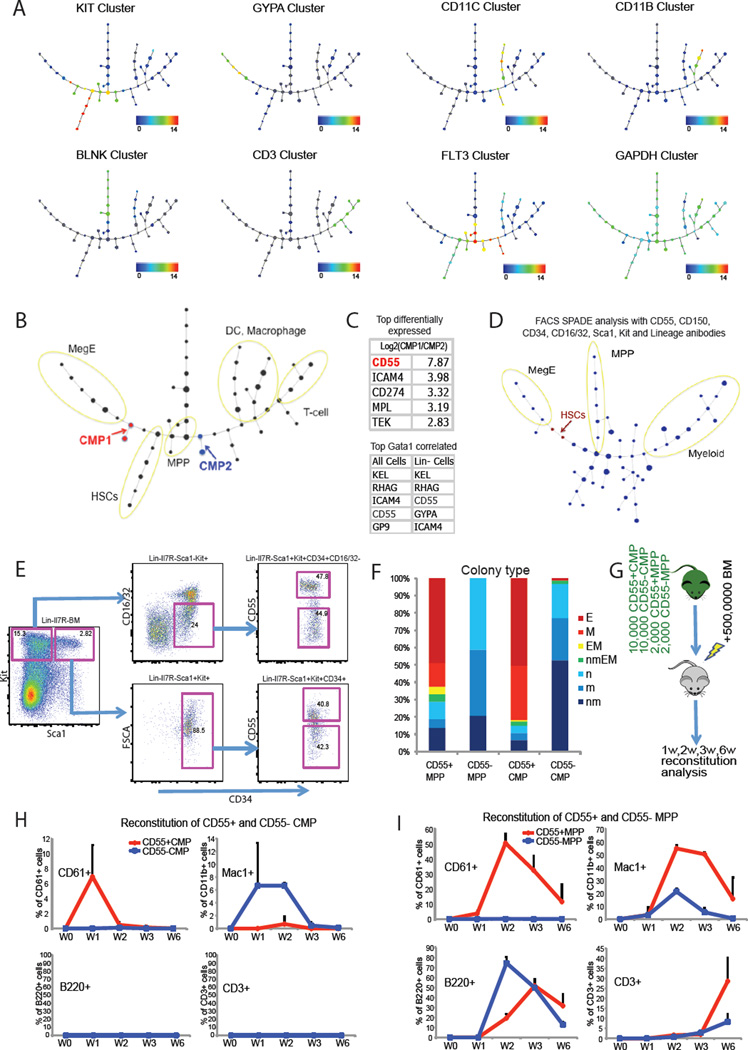
We hypothesized that the similarity of different single cell signatures and continuity of transitional states during differentiation could form the foundation of an in silico strategy to organize high-dimensional data into ordered, stepwise cell fate commitment pathways. To accomplish this, we first removed redundancy by extracting the average value of 40 distinct gene expression clusters from the entire dataset (Table S3), and then used SPADE (spanning-tree progression analysis of density-normalized events) (Bendall et al., 2011; Qiu et al., 2011) analysis to distill 40 dimensional single cell data down to a single interconnected cluster of transitional cell populations. The unsupervised computationally constructed hierarchy shows high resemblance to the hematopoietic differentiation lineage tree (Figure 4A). Different cell lineages are readily separated into distinct branches, as revealed by the overlaid expression level of different gene clusters. Branches expressing Kit cluster and Gypa cluster genes correspond to stem and progenitor, and to MegE lineage cells, respectively. The dendritic, macrophage, B cell, T cell branches, as well as lymphomyeloid progenitor cells, are marked by expression of CD11c, CD11b, Blnk, CD3 and Flt3 clusters, respectively. The Gapdh endogenous control cluster is expressed broadly.
Figure 4. Mapping cellular hierarchy by lineage progression analysis.
(A) Spanning-tree progression analysis of density-normalized events from single cell expression pattern of 40 gene clusters. The information regarding each clusters is listed in Table S3. Overlaid expression pattern of different gene clusters helped to define distinct cell lineages. Color scale is as described in Figure 1B. (B) Single cell SPADE hierarchy suggests early separation of MegE lineage and lymphomyeloid lineage. (C) CD55 is the most differentially expressed MegE cell surface marker between the CMP1 and CMP2. In addition, CD55 is highly correlated with Gata1 expression across the single cell data set. (D) SPADE analysis of FACS data from mouse bone marrow stained with CD55, CD150, CD34, CD16/32, Sca1, Kit and lineage antibodies. Only Lin-Kit+ (as defined by Lineage Signal <1000, Kit Signal>1000) data points are included in the analysis to reduce complexity. The two cell cluster nodes that contain most of the HSCs (as defined by CD150 Signal > 1000, CD48 Signal < 1000, CD34 Signal < 1000, Sca1 Signal > 1000) containing nodes are labeled in red. The two nodes are closely related with MegE branch as defined by overlaid expression in Figure S4C. (E) CD55 can be used to separate both CMP and MPP progenitors. (F) In vitro colony forming assays using EPO containing methylcellulose suggest that CD55 divides both CMP and MPP into functionally different subpopulations. 150 cells from each population were plated in 1.5mL of Methocult M3434 (Stem Cell Technologies) in duplicates. E: Erythrocytes; M: Megakaryocytes; m: monocytes; n: neutrophils. (G)-(I) Reconstitution experiment using Actβ GFP mice validated the early separation of MegE lineage potential and lymphomyeloid potential in vivo. X-axes corresponds to sampling time. Y-axes corresponds to GFP% of the total reconstituted cells. Mice are irradiated by two doses of 5 Gy with a 4 hours interval. Data are represented as mean of 4 biological replicates for each group. Error Bars correspond to standard deviation. See also Figure S4.
In the hierarchy generated from single cell expression data, the MegE lineage branch is closely connected to the long-term repopulating HSC branch. These data suggest that the MegE lineage separates very early from lymphomyeloid lineage cells. Upon inspection of the composition of different nodes, we found that phenotypic CMP cells are located on two separate differentiation pathways, with half merged to the MegE lineage and half merged to the lymphomyeloid lineage (Figure 4B). This pattern is inconsistent with the conventionally portrayed, classical differentiation scheme that positions the MegE progenitor after the bifurcation of CMP and CLP, and is reminiscent of an alternative model (Adolfsson et al., 2005; Pronk et al., 2007).
To validate this alternate scheme functionally, we sought to predict an early MegE lineage specific marker from our data resource. We compared gene expression differences between the two separated CMP compartments (CMP1 and CMP2), and identified CD55 as the most differentially expressed MegE marker (Figure 4C). In addition, we found that CD55 expression strongly correlated with the Gata1 transcription factor (Figure 4C and S4A), a master regulator of MegE lineage specification (Arinobu et al., 2007; Fujiwara et al., 1996; Iwasaki et al., 2003). FACS analyses indicate that Lin-Kit+Sca1- cells can be separated by CD55 into two main compartments (Figure S4B). To overcome the limitation of traditional 2D gating strategy, we used SPADE analysis to analyze multi-dimensional FACS data from mouse bone marrow stained with CD55, CD150, CD34, CD16/32, Sca1, Kit and lineage antibodies. We focused on Lin- Kit+ data points and generated a simplified lineage tree with 7 dimensional single cell profiles. Consistent with our qPCR expression findings, the MegE lineage branch is closely connected with the HSC containing cell cluster nodes (Figure 4D and S4C), confirming early MegE specification.
We next separated CMP (Lin-IL7R-Sca1+Kit+CD34+CD16/32lo) and MPP (Lin-Sca1+Kit+CD34+) compartments into CD55+ and CD55- subpopulations (Figure 4E) and tested their function using in vitro colony forming assays. Both CD55+ MPP and CD55+ CMP produce predominantly erythroid and megakaryocytic colonies, whereas few MegE colonies arise from CD55- MPP or CD55- CMP, revealing a functional difference in these early progenitor compartments (Figure 4F and S4D). In order to confirm the early MegE separation in vivo, we used Actβ-GFP mice for transplantation studies (Figure 4G). CD55+ CMPs transiently give rise to CD61+ platelets, whereas CD55- CMPs produce mainly myeloid cells (Figure 4H). CD55+ MPPs achieved more than 50% platelet reconstitution, whereas there was no reproducible contribution of CD55-MPPs to CD61+ platelets (Figure 4I and S4E). Importantly, CD55- CMPs and CD55- MPPs failed to produce platelets in vivo, whereas CD150- progenitors exhibited robust MegE potential (Pronk et al., 2007), suggesting that CD55 is an improved marker for separating early MegE progenitors. In conclusion, by computational analysis of single cell data, we have predicted and validated CD55 as a marker to establish a functional separation between early MegE and lymphomyeloid differentiation at both CMP and MPP stages.
Genetic network construction by single cell analysis
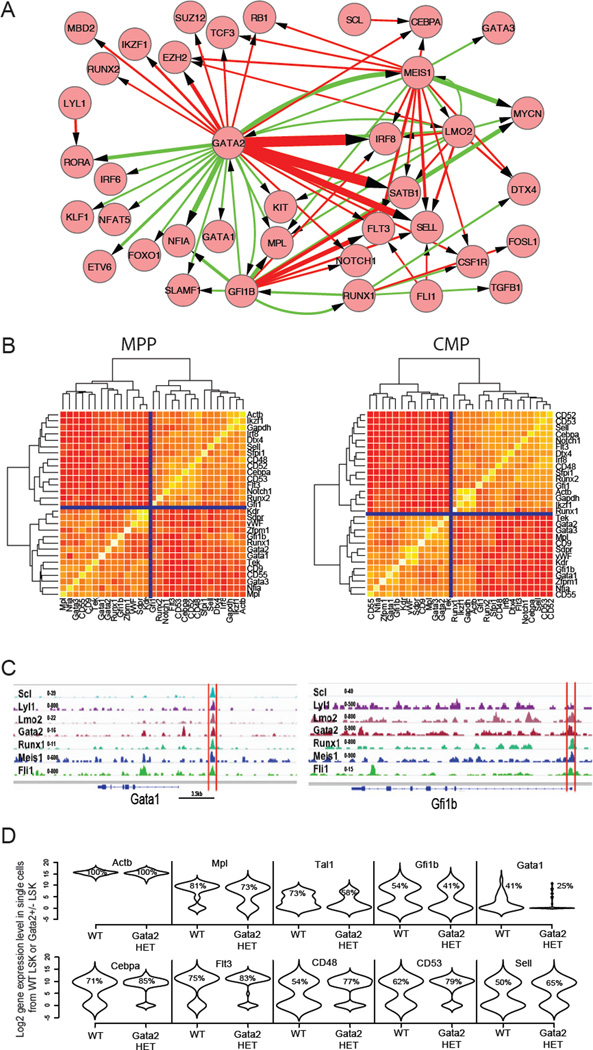
To explore potential molecular mechanisms underlying early hematopoietic lineage specification, we designed primers to assay expression of an additional 180 genes, including lineage specific transcription factors, epigenetic modifiers, and cell cycle regulators. We assayed single cells from HSCs (CD48-CD34-CD150+LSK), MPP (CD34+LSK), CMP, MEP, GMP and CLP populations (Table S4), and calculated gene expression covariance across the data set to uncover hidden regulatory links. We then used Cytoscape software to integrate expression correlations with published ChIP-seq binding datasets for ten major stem cell transcriptional regulators (including Scl/Tal1, Lyl1, Lmo2, Gata2, Runx1, Meis1, PU.1, Erg, Fli1 and Gfi1b) from HPC-7 cell line (Wilson et al., 2010). The network (Figure S5A) only depicts links in which the covariance was above 0.1 for correlated genes (green edges) or below −0.1 for anticorrelated genes (red edges). The network contains 76 nodes, connected through 71 edges between correlated genes and 74 edges between anticorrelated genes. Figure 5A highlights the transcription factor components of the complete network. As revealed, Gata2, a central hematopoietic stem cell regulator (Tsai et al., 1994; Wilson et al., 2010), lies at the core of the lineage specification pathway (Figure 5A), and positively correlates with a MegE lineage module (characterized by Gata1, Gfi1b, Nfia and Klf1), and negatively correlates with a lymphomyeloid module (characterized by Flt3, Sell, Cebpa and Notch1). This is also depicted on a gene-to-gene correlation heatmap in Figure 5B. Time course single cell tracing experiments suggest that up-regulation or down-regulation of Gata2 marks the first molecular event during colony formation (Figure S5B). The correlation in expression level between Gata2 and Gata1 is maintained during both in vivo and in vitro differentiation (Figure S5C). As revealed in Figure 5C, Gata2, Runx1, Meis1, Scl, Lyl1 and Lmo2 co-occupy at Gata1 and Gfi1b regulatory regions (Figure 5C). The stem cell transcription factor Gata2 occupies regulatory elements of multiple MegE lineage related genes, as well as HSC enriched genes (Figure S5D).
Figure 5. Genetic regulation during HSC differentiation.
(A) A genetic network constructed by Cytoscape using transcription factor ChIP-seq binding information and single cell level gene expression correlation data. It highlights transcription factors components within the complete network in Figure S5A. Green arrow corresponds to positive correlation, while red arrow corresponds to anti correlation. The width of the line corresponds to absolute value of the covariance between two linked gene nodes. (B) Gene to gene correlation heatmaps containing HSC, MegE, Myeloid and Lymphoid modules in MPP and CMP. (C) HSC module transcription factors co-occupy Gata1 and Gfi1b upstream region. (D) Gene expression level distribution of LSK single cells from wildtype and Gata2 heterozygous mice are presented with violin density plots. The percentages of cells with positive expression levels are marked on the violin plots. Note the decrease in MegE primed cells (Gfi1b+ or Gata1+) cells and increase in lymphomyeloid primed cells (Cebpa+, Flt3+, CD53+ or Sell+) in the Gata2 +/− LSK population. See also Figure S5.
As a functional test of this predicted genetic network, we examined the consequences of perturbation of the level of Gata2. Since Gata2 −/− embryos die early due to hematopoietic failure (Tsai et al., 1994), we analyzed gene expression changes in viable Gata2+/− mice at single cell resolution. Consistent with a previous report (Rodrigues et al., 2005), we observed a reduction in the size of the LSK population in Gata2 heterozygous mice as compared with wildype. Single cell gene expression analysis of Gata2 +/+ and Gata2 +/− LSK reveals that haploinsuffciency is associated with an altered regulatory network during early lineage differentiation (Figure 5D and S5E), revealing sensitivity of the network to modest quantitative changes in Gata2 expression. Haploinsufficiency for Gata2 leads to down regulation of the MegE marker Gfi1b and Gata1, and up regulation of lymphomyeloid markers, including Flt3, Sell, CD34, CD53 and Cebpa in hematopoietic stem and progenitor cell populations (Figure 5D and S5E). Taken together, single cell level gene expression in combination with functional studies validates a genetic network underlying early differentiation of MegE lineage from the lymphomyeloid lineage.
MegE priming in the most primitive HSCs
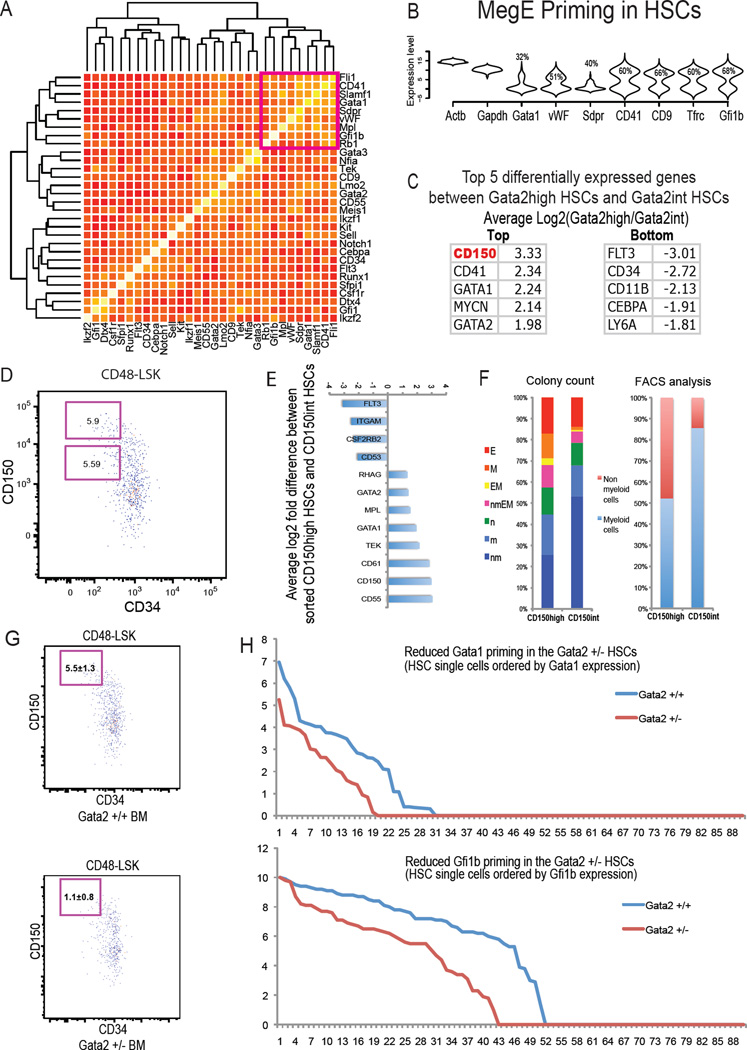
In single cell in vitro tracing experiments (Figure S5B), we noticed that pure megakaryocytic colonies are the first to emerge in cultures of HSCs. These results encouraged us to investigate the heterogeneity and existing MegE network within the most primitive HSC (CD48-CD34-CD150+LSK) population. Remarkably, a MegE module, characterized by expression of Fli1, CD41, CD150, Gata1, vWF, Mpl and Gfi1b, maintains high correlation in single HSCs (Figure 6A). MegE lineage specific gene expression is detected in HSCs purified by different enrichment protocols, and further confirmed by single cell NanoString technology (Figure 6B; Figure S6A and S6B; Table S5). Such transcriptional priming does not appear to be stochastic, but rather controlled by an intertwined HSC regulatory network. We ranked single HSCs by the expression level of Gata2 and compared gene expression between _Gata2_high HSCs (top 50%) and _Gata2_int HSCs (bottom 50%). CD150 emerged as a candidate marker for separating HSCs according to different levels of Gata2 expression (Figure 6C), as well as different degrees of MegE priming (Figure S6C). To confirm these differences, we FACS sorted HSCs into CD150high and CD150int compartments for gene expression analysis (Figure 6D). Indeed, CD150high HSCs express higher levels of Gata2, Gata1, CD61, as well as other MegE lineage related genes (Figure 6E). In colony forming assays, CD150high HSCs generate greater numbers of MegE lineage containing colonies than CD150int HSCs (Figure 6F). Similar biased differentiation readouts were also seen, in HSCs that were separated by relative expression levels of CD55, CD41 or CD9 (Figure S6D). These results suggest that MegE differentiation bias is already established at the HSC level.
Figure 6. MegE lineage priming in HSCs.
(A) Gene to gene correlation heatmaps reveals correlation of MegE lineage markers in single cells from HSCs (CD48-CD34-CD150+LSK). (B) Violin plot suggests significant MegE lineage priming in HSCs (CD48-CD34-CD150+LSK). (C) CD150 stands out as the top differentially expressed gene between Gata2high HSCs and Gata2int HSCs. (D) FACS sorting of CD150high and CD150int HSCs. (E) Gene expression difference between the sorted CD150high versus CD150int HSCs. (F) In vitro colony forming assays using Methocult M3434 (Stem Cell Technologies) suggest that CD150high HSCs produce more MegE lineage containing colonies than the CD150int HSCs. FACS analysis of day7 methylcellulose cultures also suggests a decreased percentage of CD11b+ or Gr1+ myeloid cells generated from the CD150high HSCs when compared to CD150int HSCs. CD11b- and Gr1- cells were defined as non-myeloid cells. (G) Gata2 haploinsufficiency results in a reduction of CD150high HSCs. Three animals were analyzed for each genotype; results are shown as mean±SD. (H) Gata2 haploinsufficency results in a reduction of Gata1 and Gfi1b priming in the HSC compartment. A total of 87 single cells were analyzed for each genotype. Single cells are ordered by Gata1 or Gfi1b expression. See also Figure S6.
The positive correlation of Gata2 with the MegE priming expression suggests that the regulatory network within HSCs is intrinsically unstable. As such, higher levels of Gata2 in HSCs may activate MegE lineage expression and promote MegE lineage skewing. When stained with the full panel of HSC markers, we observed a reduced number of CD150high HSCs in the Gata2 haploinsufficient state (Figure 6G). In addition, in the most primitive HSCs of Gata2 +/− mice, we observed a reduction in the number _Gata1_+ or _Gfi1b_+ HSCs, as well as the average level of MegE priming (Figure 6H). Consistent with these findings, overexpression of Gata2 has been reported to promote MegE differentiation (Huang et al., 2009; Kitajima et al., 2006). The characterization of MegE priming in HSCs supports the cellular hierarchy and genetic network derived from single cell expression data, and illustrates the power of single cell analysis in detecting the earliest regulatory events during stem cell differentiation.
Single cell analysis of the AML cellular hierarchy
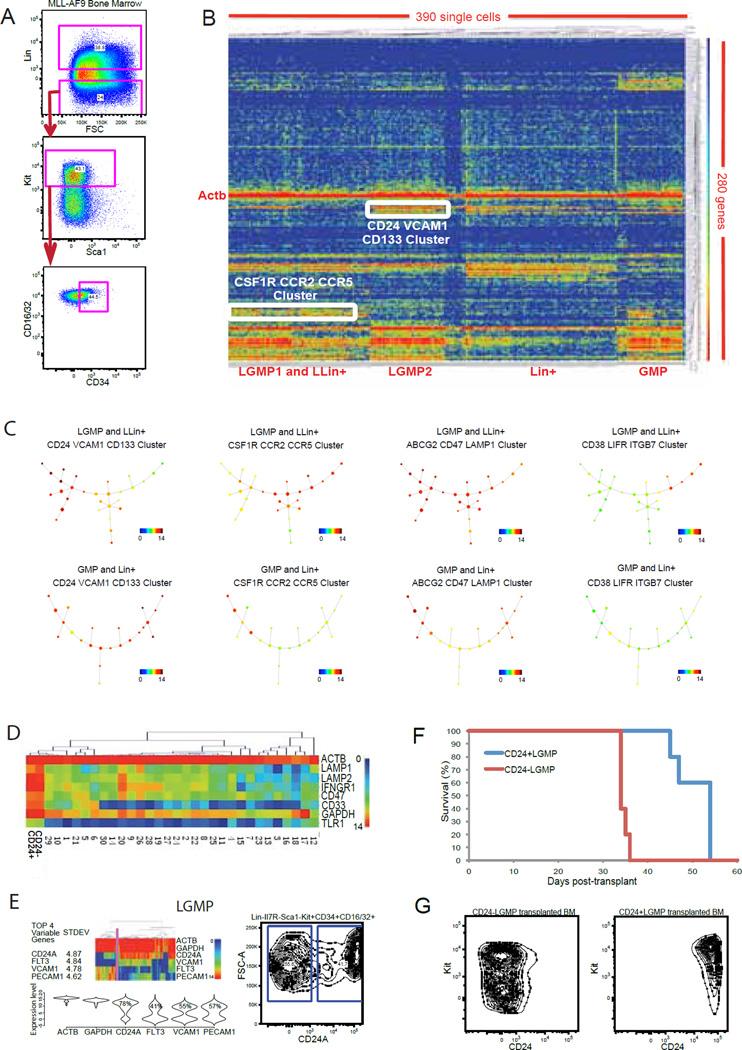
Having obtained a comprehensive data set in the wild-type hematopoietic system, we next applied the single cell expression approach to characterization of leukemic stem cells (LSCs) in MLL-AF9 driven AML, a clinically relevant model of hematopoietic malignancy (Krivtsov et al., 2006; Neff et al., 2012). In this model, LSCs resemble GMPs, and are hence described as LGMPs. Others, however, have described alternative cellular hierarchies of AML (Gibbs et al., 2012; Somervaille and Cleary, 2006). We generated MLL-AF9 primary leukemia in mice (Neff et al., 2012), and profiled single cells of the originally defined LGMP LSC population (Lin-Il7r-Kit+Sca1-CD34+CD16/32+), as well as the Leukemic Lin+ (LLin+) population from bone marrow (Figure 7A and Table S1). As shown in Figure 7B, hierarchical clustering of gene expression data from leukemia cells and the wildtype myeloid cells reveals clear separation of the two groups. The Lin+ leukemia population (LLin+) clusters closely with a group of LGMP cells, suggesting that lineage marker expression does not define a clear hierarchy in the leukemia. Two strong gene clusters are observed in the leukemia cells: a Csf1r, Ccr2, Ccr5 cluster; and a CD24, Vcam1, CD133 cluster (Figure 7B). We adapted SPADE to analyze the data (Figure 7C). To allow for comparison of the wildtype and leukemia lineages, we extracted 40 clusters from the combined data sets of LGMP, LLin+, GMP and Lin+ single cells (Table S6). We then used these clusters to infer lineage hierarchy for both cellular systems. From the overlaid expression level of different gene clusters, we observed clear separation of the _CD24_+ lineage branch and the _Csf1r_+ lineage branch within the tested leukemia cells (Figure 7C). By comparing the two main leukemia cell type signatures with other hematopoietic cell types, we find that MLL-AF9 leukemia cells display a unique signature with high expression of Lamp1, Lamp2, Ifngr1, CD47 and CD33 (Figure 7D). Notably, the leukemia cellular state differs from other hematopoietic cellular states both at the single cell and population levels.
Figure 7. Single cell analysis of the AML cellular hierarchy.
(A) Single cell sorting strategy for different leukemia compartments in the MLL-AF9 AML mouse model. (B) A heatmap showing hierarchical clustering of gene expression signatures from 390 single cells from wildtype myeloid cells (Lin-Il7r-Kit+Sca1- CD34+CD16/32+ or Lin+ bone marrow) and MLL-AF9 primary leukemia cells (Lin-Il7r-Kit+Sca1-CD34+CD16/32+ or Lin+ bone marrow). Each row corresponds to a specific gene; each column corresponds to a particular single cell. White boxes highlight strongly correlated gene and cell clusters. Color scale is as described in Figure 1B. (C) SPADE analysis of the wildtype myeloid hierarchy and leukemia hierarchy using the high dimensional single cell data. Overlaid expression pattern of different gene clusters helped to define distinct cell lineages in the MLL-AF9 leukemia system. (D) Gene expression clustering heatmap of gene expression from the main leukemia cell clusters and all the main hematopoietic cell clusters reveals distinct leukemia specific expression. (E) Dissection of heterogeneity in the LGMP (Lin-Il7r-Kit+Sca1-CD34+CD16/32+) according to described method in Figure 3. (F) Survival of secondary recipient mice receiving 800 CD24+ LGMP or CD24-LGMP cells. (G) Reconstitution of two leukemia lineages in the secondary recipient bone marrow. See also Figure S7.
In the previously defined LGMP population in which LSCs are highly enriched (Krivtsov et al., 2006), we observed clear heterogeneity. Guided from single cell data, we separated the LGMP into two populations using CD24 antibody (Figure 7E). To assess potential functional difference of these two compartments, we transplanted each into sub-lethally irradiated secondary recipients. Both CD24-LGMP and CD24+LGMP are capable of initiating AML (Figure 7F). However, mice transplanted with CD24+LGMPs exhibited a marked delay in disease progression. Analysis of the bone marrow from secondary leukemia mice indicated that CD24− leukemia cells and CD24+ leukemia cells maintain their respective signatures and fail to reconstitute each other during clonal expansion (Figure 7G). Thus, CD24 marks two distinct, self-renewing clones within MLL-AF9 driven AML. Further profiling of additional intracellular regulators reveals different genetic programs used by CD24-LGMP and CD24+LGMP (Figure S7A). Interestingly, Ezh2, a core polycomb repressive complex 2 (PRC2) component, is overexpressed in CD24− LGMPs (Figure S7A). Our analysis also reveals high variation of Ezh2 at the single cell level, which strongly correlates with Ccna2, Ccnb1 and Ccnb2 expression (Figure S7B). Such correlation may account in part for the more aggressive behavior of the CD24− Ezh2high leukemia clone, as compared with the CD24+ Ezh2low leukemia clone. In microarray data of synchronized Hela cells (Whitfield et al., 2002), Ezh2 expression is lowest in G1 and peaks at S phase (Figure S7C). In addition, many cell cycle regulators are direct targets of PRC2, as assessed from PRC2 chromatin occupancy data (Figure S7D–S7G). Moreover, inhibition of Ezh2 function with the specific inhibitor GSK126 (McCabe et al., 2012) leads to an increase in G1 phase cells and a decrease in S phase cells in MLL-AF9 cultures (Figure S7H). Our findings are in general agreement with the observation that EZH2 overexpression correlates with poor prognosis in several tumor types (Cavalli, 2012; McCabe et al., 2012).
DISCUSSION
Single cell analysis technologies provide a powerful approach to the study of rare cell types and cell heterogeneity. For both genome analysis and transcriptome analysis of single cells, amplification of small amounts of material is required and presents technical challenges. For assessment of gene expression, single cell high throughput qPCR has several advantages. First, it utilizes one tube, one-step single cell sequence specific preamplification, which involves minimal sample handling time and allows for high throughput cDNA library generation. Second, the targeted PCR approach enables specific amplification of lowly expressed genes. Finally, Biomark (Fluidigm) microfluidic qPCR permits well-controlled, parallel analysis of 96 single cell samples. The system minimizes technical variation, allowing for comparison of different samples without normalization. A major challenge relates to primer dimer formation during multiplexed sequence specific preamplification step, which generates false positive signals (Guo et al., 2010). To overcome this obstacle, we have introduced multiplexed primer design, lowered the preamplification primer concentration, and included nested primers to avoid primer dimer signals. These optimizations significantly increased the throughput of single cell qPCR technology, and permitted analysis of the cell surface repertoire in many single cells from a broad range of tissue types. We show with functional validation that such data sets can be used for classification of cell type, dissection of heterogeneity, mapping of cellular hierarchy and computational construction of genetic networks.
We have applied our analysis to examine cellular lineages of the mouse hematopoietic system. Computational lineage progression analysis provided an unbiased view of cellular state transition during differentiation from HSCs. Our findings independently support an alternative hematopoietic hierarchy first proposed by Jacobsen and colleagues (Adolfsson et al., 2005) and provide a molecular model for early MegE lineage separation. The sensitivity of the assay allowed detection of coordinated MegE transcriptional priming within the most highly enriched HSCs. We then extended the analysis to a robust, clinically relevant model of AML. At the level of single cells, we showed that leukemia cells are intrinsically distinct from any of the wildtype hematopoietic lineages. Interestingly, the most significant heterogeneity within the leukemia corresponded to two independent, disease-initiating clones. Moreover, we found that Ezh2 is overexpressed in the more highly proliferative leukemia cells and uncovered a link with cell cycle progression.
No two cells are identical, a concept evident in our dataset. Even the most closely related cells, which correspond to two erythroid progenitor cells from our complete data clustering heatmap (Figure 2B), exhibit differences in gene expression patterns. Variation detected in single cell gene expression data may reflect biological and/or technical noise, or may correspond to function. Distinguishing these two types of variation is very important. Here, we assayed many single cell samples and searched for correlated gene clusters rather than variation in expression of individual genes. Such correlated gene expression behavior is more likely to represent genetic network function rather than biological noise. By this approach we correlated MegE priming in the HSCs and correlated Ezh2 dynamics with cell cycle, which were then both functionally validated. Single cell gene expression data is extremely valuable for extracting such correlated gene expression clusters, because single cells represent the fundamental unit of genetic network regulation.
The mammalian epigenetic landscape contains numerous transitional cellular states within lineage differentiation pathways. Comprehensive mapping of this landscape requires single cell gene expression analysis in order to represent all possible states. Such an assay needs to be both quantitative and thorough, so that the data are experimentally robust and reflects all major cell types. We suggest that the strategy described here satisfies these requisites. In order to validate biological differences in cell populations, we have relied extensively on study of cell surface markers, as available antibodies can then be used for prospective cell isolation. The approach is readily applicable to other biological contexts. Its use should facilitate identification of new surface markers for functional assessment of stem and progenitor cells, and the construction of cellular hierarchies in other organ systems. The strategy is suitable for deconvoluting cellular heterogeneity within different types of cancers. Further accumulation of data sets from diverse contexts should eventually allow for the mapping of all non-redundant cellular states on the mammalian differentiation hierarchy.
EXPERIMENTAL PROCEDURES
Multiplexed primer design for single cell analysis
Gene symbol list for commonly used surface markers is summarized from two resources: a comprehensive mouse cell surface antigens review paper (Lai et al., 1998), and the eBioscience website mouse cellular antigen charts (http://www.ebioscience.com/resources/mouse-cd-chart.htm). Gene symbols are then converted to mRNA refseq ID by DAVID tools (http://david.abcc.ncifcrf.gov/). mRNA sequences for each genes are retrieved from USCS table browser; only common regions are used for genes with different isoforms. Multiplexed gene specific primers are designed using a Primer3 (http://primer3.wi.mit.edu/) based algorithm to ensure that each primer within the designed pool has a maximum complimentary sequence of 7 bp to all the other primers. All primers (Table S7) are synthesized and provided by Boston Open Labs (http://bolresearch.com/).
FACS sorting and single cell collection
Seven to twelve-week old C57Bl/6 mice or Actβ-GFP C57Bl/6 transgenic mice were used throughout this study (expect for fetal liver and aged mice). Bone marrow cells were isolated by crushing iliac crest bones, femurae and tibiae in PBS containing 5% FCS and 2mM EDTA. After red blood cell lysis, the remaining cells were stained with monoclonal antibodies, analyzed and sorted on the BD FACSAria II (BD Bioscience). Individual cells were sorted directly into 96 well PCR plates loaded with PCR buffer under single cell mode. Monoclonal antibodies and conjugations used in this study are found in Table S7. All data were analyzed with FlowJo (Tree Star).
One tube single cell sequence specific preamplification
Individual primer sets (total of 300) were pooled to a final concentration of 0.1µM for each primer. Individual cells were sorted directly into 96 well PCR plates loaded with 5µL RT-PCR master mix (2.5µL CellsDirect reaction mix, Invitrogen; 0.5µL primer pool; 0.1µL RT/Taq enzyme, Invitrogen; 1.9µL nuclease free water) in each well. Sorted plates were immediately frozen on dry ice. After brief centrifugation at 4°C, the plates were immediately placed on PCR machine. Cell lyses and sequence-specific reverse transcription were performed at 50°C for 60 minutes. Then reverse transcriptase inactivation and Taq polymerase activation was achieved by heating to 95°C for 3 min. Subsequently, in the same tube, cDNA went through 20 cycles of sequence-specific amplification by denaturing at 95°C for 15 sec, annealing and elongation at 60°C for 15 min. After preamplification, PCR plates were stored at −80°C to avoid evaporation.
High throughput microfluidic realtime PCR
Pre-amplified products were diluted 5-fold prior to analysis. Amplified single cell samples were analyzed with Universal PCR Master Mix (Applied Biosystems), EvaGreen Binding Dye (Biotium) and individual qPCR primers using 96.96 Dynamic Arrays on a BioMark System (Fluidigm). 3 Dynamic arrays loaded with different primer sets were used for each sample plate. Ct values were calculated using the BioMark Real-Time PCR Analysis software (Fluidigm).
Single cell NanoString
Reporter probes are designed and synthesized by NanoString R&D team. Target sequences are amplified from single cells using one tube single cell sequence specific preamplification as describe before. 25% of the amplified cDNA are subject to gene expression quantification using the GEN2 Digital Analyzer. Raw counts are compiled, normalized and analyzed using nSolver. The data are then subtracted with the background signal and transformed to Log2 scale before analysis.
Computational processing of single cell data
A background Ct of 28 was used for all realtime signals. Samples with low Actb expression level (Ct higher than 18) are outliers of normal distribution and are excluded from the analysis. These samples had low or no expression for all the other genes, suggesting that they correspond to empty wells or bad single cell samples. Hierarchical clustering was done with MultiExperiment Viewer (MeV) program. For all hierarchical clustering heatmaps, the rainbow scheme color scale is set from 0 to 14 corresponding to Log2 gene expression above background of 28. GEDI plots are generated using the gene expression dynamics inspector. Each pixel on the 10.10 GEDI map corresponds to a particular mini gene cluster generated by the software. Violin plot, box plot and correlation heatmap were generated with R software. SPADE analysis was performed with Matlab. Lineage specific gene lists for the 180 intracellular regulator assay set and for Figure S5D are generated from the Immgen website analysis tool. ChIP-seq peak visualization was done with IGV program. The genetic networks in Figure 5A and S5A were constructed using Cytoscape 3 software.
Supplementary Material
01
02
03
04
05
06
07
08
Highlights.
- Robust methodology for single cell analysis of the cell surface repertoire
- Comprehensive single cell analysis for the mouse hematopoietic system
- Lineage progression analysis and network construction using single cell data
- Characterization of the unique cellular hierarchy in a mouse AML model
ACKNOWLEDGEMENTS
We thank H. Skaletsky from Whitehead Institute for extensive help on the multiplexed primer design. Y. Fujiwara, E. Baena, O. Yilmaz, M. Nguyen, X. Han, V. Bragt, D. Linn and J. Buchman for help with different parts of the sample preparations. H. Huang, Z. Li, D. Scadden, H. Xie, and H. Hock for insightful discussions on the project. This work was supported by funding from NIH and the Harvard Stem Cell Institute (S.H.O). S.H.O. is an investigator of the Howard Hughes Medical Institute (HHMI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
REFERENCES
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G. Molecular biology. EZH2 goes solo. Science. 2012;338:1430–1431. doi: 10.1126/science.1232332. [DOI] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs KD, Jr, Jager A, Crespo O, Goltsev Y, Trejo A, Richard CE, Nolan GP. Decoupling of tumor-initiating activity from stable immunophenotype in HoxA9-Meis1-driven AML. Cell Stem Cell. 2012;10:210–217. doi: 10.1016/j.stem.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Huang Z, Dore LC, Li Z, Orkin SH, Feng G, Lin S, Crispino JD. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol Cell Biol. 2009;29:5168–5180. doi: 10.1128/MCB.00482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Highly multiplexed and strand-specific single-cell RNA 5' end sequencing. Nat Protoc. 2012;7:813–828. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Wells RA, Cantor AB, Watanabe S, Akashi K. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 2003;19:451–462. doi: 10.1016/s1074-7613(03)00242-5. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kitajima K, Tanaka M, Zheng J, Yen H, Sato A, Sugiyama D, Umehara H, Sakai E, Nakano T. Redirecting differentiation of hematopoietic progenitors by a transcription factor, GATA-2. Blood. 2006;107:1857–1863. doi: 10.1182/blood-2005-06-2527. [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Lai L, Alaverdi N, Maltais L, Morse HC., 3rd Mouse cell surface antigens: nomenclature and immunophenotyping. J Immunol. 1998;160:3861–3868. [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Moignard V, Macaulay IC, Swiers G, Buettner F, Schutte J, Calero-Nieto FJ, Kinston S, Joshi A, Hannah R, Theis FJ, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol. 2013;15:363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a lineage of multipotent hematopoietic progenitors. Development. 1997;124:1929–1939. doi: 10.1242/dev.124.10.1929. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Muller-Sieburg CE, Whitlock CA, Weissman IL. Isolation of two early B lymphocyte progenitors from mouse marrow: a committed pre-pre-B cell and a clonogenic Thy-1-lo hematopoietic stem cell. Cell. 1986;44:653–662. doi: 10.1016/0092-8674(86)90274-6. [DOI] [PubMed] [Google Scholar]
- Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, Xie H, Orkin SH, Armstrong SA. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci U S A. 2012;109:5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, Enver T, Vyas P, Scadden DT. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Visser JW, Bauman JG, Mulder AH, Eliason JF, de Leeuw AM. Isolation of murine pluripotent hemopoietic stem cells. J Exp Med. 1984;159:1576–1590. doi: 10.1084/jem.159.6.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06
07
08