Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells (original) (raw)
. Author manuscript; available in PMC: 2014 Jun 1.
Published in final edited form as: Nat Biotechnol. 2013 Nov 10;31(12):1126–1132. doi: 10.1038/nbt.2720
Abstract
Genome sequencing of single cells has a variety of applications, including characterizing difficult-to-culture microorganisms and identifying somatic mutations in single cells from mammalian tissues. A major hurdle in this process is the bias in amplifying the genetic material from a single cell, a procedure known as polymerase cloning. Here we describe the microwell displacement amplification system (MIDAS), a massively parallel polymerase cloning method in which single cells are randomly distributed into hundreds to thousands of nanoliter wells and simultaneously amplified for shotgun sequencing. MIDAS reduces amplification bias because polymerase cloning occurs in physically separated nanoliter-scale reactors, facilitating the de novo assembly of near-complete microbial genomes from single E. coli cells. In addition, MIDAS allowed us to detect single-copy number changes in primary human adult neurons at 1–2 Mb resolution. MIDAS will further the characterization of genomic diversity in many heterogeneous cell populations.
The genetic material in a single cell can be amplified in vitro by DNA polymerase into many clonal copies, which can then be characterized by shotgun sequencing. Single-cell genome sequencing has been successfully demonstrated on microbial and mammalian cells1–6, and applied to the characterization of the diversity of microbial genomes in the ocean7, somatic mutations in cancers8, 9 and meiotic recombination and mutation in sperm3, 10. The most commonly used method for amplifying DNA from single cells is multiple displacement amplification (MDA)2. Currently, the major technical challenge in using MDA is the highly uneven amplification of the one or two copies of each chromosome in a single cell. This high amplification bias leads to difficulties in assembling microbial genomes de novo and inaccurate identification of copy number variants (CNV) or heterozygous single nucleotide changes in single mammalian cells. Recent developments of bias-tolerant algorithms11, 12 have greatly mitigated the effects of uneven read depth on de novo genome assembly and CNV calling, yet an unusually high sequencing depth is still required, making this approach impractical for organisms with large genomes.
Several strategies have been developed to reduce amplification bias, including reducing the reaction volume13, 14 and supplementing amplification reactions with single-strand binding proteins or trehalose5, 15. Post-amplification normalization by digesting highly abundant sequences with a duplex-specific nuclease has also been used to markedly reduce bias2. Despite these efforts, amplification bias still remains the primary technical challenge in single-cell genome sequencing. A relatively large amount of sequencing is still necessary to obtain a high-quality genome sequence even with these improvements. Using cells that contain multiple copies of the genome or multiple clonal cells has been the only viable solution to achieve near complete genome coverage with MDA16, 17. Other methods such as MALBAC utilize quasi-linear amplification to reduce exponential amplification bias18; however, the specific polymerase required can introduce a higher level of amplification error, complicating further analysis.
We reasoned that whole-genome amplification is always prone to bias because repeated priming in similar locations becomes exponentially more favorable as the reaction continues. Thus, we hypothesized that bias could be reduced by limiting the reaction so that just enough amplification occurs to allow sequencing, thereby limiting the potential iterations of repeated priming. In addition, we supposed that reducing the reaction volume by ~1,000 fold to nanoliter levels, which increases the effective concentration of the template genome, might both reduce contamination and improve amplification uniformity, as the higher concentration of template would lead to more favorable primer annealing kinetics in the initial stages of MDA13, 14.
To test these hypotheses, we developed the microwell displacement amplification system (MIDAS), an approach that allows for highly parallel polymerase cloning of single cells in thousands of nanoliter reactors. Each reactor spatially confines a reaction within a 12 nL volume, to our knowledge the smallest volume that has been implemented to date. Coupled with a low-input library construction method, we achieved highly uniform coverage in the genomes of both microbial and mammalian cells. We demonstrated substantial improvement both in de novo genome assembly from single microbial cells and in the ability to detect small somatic copy number variants in individual human adult neurons with minimal sequencing effort.
RESULTS
MIDAS implements massively parallel polymerase cloning
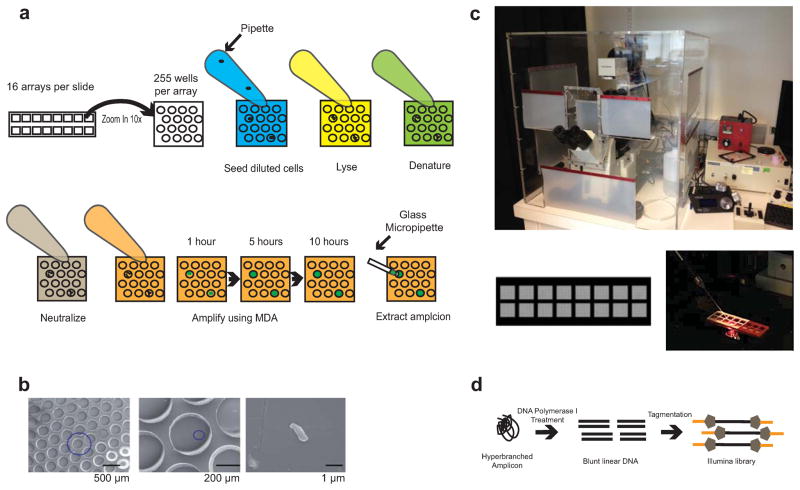
We designed and fabricated microwell arrays of a size comparable to standard microscope slides. The format of the arrays, including well size, pattern and spacing, was optimized to achieve efficient cell loading, optimal amplification yield and convenient DNA extraction. Each slide consisted of 16 arrays, each containing 255 microwells of 400 μm in diameter, allowing for parallel amplification of 16 separate heterogeneous cell populations (Fig. 1a). All liquid handling procedures (cell seeding, lysis, DNA denaturation, neutralization and addition of amplification master mix) required one pump of a pipette per step per array, minimizing the labor required for hundreds of amplification reactions. This system requires less of each amplification and library construction reagent than conventional methods, as each microwell spatially confines the reaction to 12 nL in volume.
Figure 1.
Microwell displacement amplification system. (a) Each slide contains 16 arrays of 255 microwells each. Cells, lysis solution, denaturing buffer, neutralization buffer and MDA master mix were each added to the microwells with a single pipette pump. Amplicon growth was then visualized with a fluorescent microscope using a real-time MDA system. Microwells showing increasing fluorescence over time were positive amplicons. The amplicons were extracted with fine glass pipettes attached to a micromanipulation system. (b) Scanning electron microscopy of a single E. coli cell displayed at different magnifications. This particular well contains only one cell, and most wells observed also contained no more than one cell. (c) A custom microscope incubation chamber was used for real time MDA. The chamber was temperature and humidity controlled to mitigate evaporation of reagents. Additionally, it prevented contamination during amplicon extraction by self-containing the micromanipulation system. An image of the entire microwell array is also shown, as well as a micropipette probing a well. (d) Complex three-dimensional MDA amplicons were reduced to linear DNA using DNA polymerase I and Ampligase. This process substantially improved the complexity of the library during sequencing.
We tested multiple cell-loading densities to ensure that each well would contain only one single cell, and initially loaded the microwells at densities of roughly 1 cell per well and 1 cell per 10 wells. By the Poisson distribution, in the 1 cell per well case, 63% should have at least one cell, but 26% could have more than one. In the 1 cell per 10 well case, no more than 0.5% of the wells should contain more than 1 cell. We confirmed that the cells were indeed being seeded at the expected distribution using fluorescent microscopy after staining cells with SYBR Green I (Supplementary Fig. 1). We thus decided to load cells at a density of 1 cell per 10 wells, ensuring that 99.5% of generated amplicons would arise from a single cell. The remaining empty wells served as internal negative controls, allowing easy detection and elimination of contaminated samples. We further confirmed proper microbial and mammalian cell seeding in microwells at the 1 cell per 10 well level by scanning electron microscopy (Fig. 1b, Supplementary Fig. 2).
After seeding of cell populations into each microwell array, we performed limited multiple displacement amplification on the seeded single cells in the partitioned microwells, each with a physically separated (save for a thin aqueous layer atop the arrays) volume of ~12 nL, in a temperature and humidity controlled chamber (Fig. 1c, Supplementary Fig. 1). We used SYBR Green I to visualize the amplicons growing using an epifluorescent microscope (Supplementary Fig. 3). A random distribution of amplicons across the arrays was observed with ~10% of the wells containing amplicons, further confirming the parallel and localized amplification within individual microwells as well as the stochastic seeding of single cells19. After amplification in the microwells, we used a micromanipulation system to extract amplicons from individual wells for sequencing (Fig. 1c). We estimated that the masses of the extracted amplicons ranged from 500 picograms to 3 nanograms.
When performing a single-cell amplification experiment, there are two potential sources of contamination that could result in an inaccurate characterization of the genome of the sample of interest. These are exogenous contamination, in which samples are exposed to cell-free DNA from environmental sources or reagents, and cross-well contamination, in which DNA from one microwell diffuses into other microwells. We ensured that neither form of contamination was occurring. To detect arrays that contained exogenous contamination, we checked for a uniform increase of fluorescent signal across all microwells. Any samples that showed this high fluorescence across all wells were removed; thus, any samples exposed to cell-free DNA were simply not analyzed. To ensure that cross-well contamination was not occurring, we performed fluorescent monitoring at 30-minute intervals during the amplification procedure. Only single wells with single amplicons originating from a single point were extracted for analysis, preventing any cross-well contamination or selection of any wells containing more than one cell (Supplementary Fig. 4). If even a miniscule amount of DNA was diffusing out of a microwell, an increased fluorescence would be observed in adjacent wells owing to amplification occurring in every well19; this diffusion was not observed in any cases. We further confirmed that cross-well contamination was not occurring by loading a mixture of human neuronal nuclei with two separate genomic backgrounds and confirming that all extracted cells corresponded only to one background (Supplementary Table 1).
To construct Illumina sequencing libraries from the extracted nanogram-scale DNA amplicons, we used a modified in-tube method based on the Nextera Tn5 transposase. Previous studies have shown that Nextera transposase-based libraries can be prepared using as little as 10 picograms of genomic DNA20. However, the standard Nextera protocol was unable to generate high-complexity libraries from MDA amplicons, resulting in poor genomic coverage (data not shown). To address this issue, we used random hexamers and DNA Polymerase I to first convert the hyperbranched amplicons into unbranched double-stranded DNA molecules, which allowed effective library construction using in vitro transposition (Fig. 1d). In addition, we used a small reaction volume to further increase the efficiency of library construction20.
Generation of a near-complete assembly from single E. coli
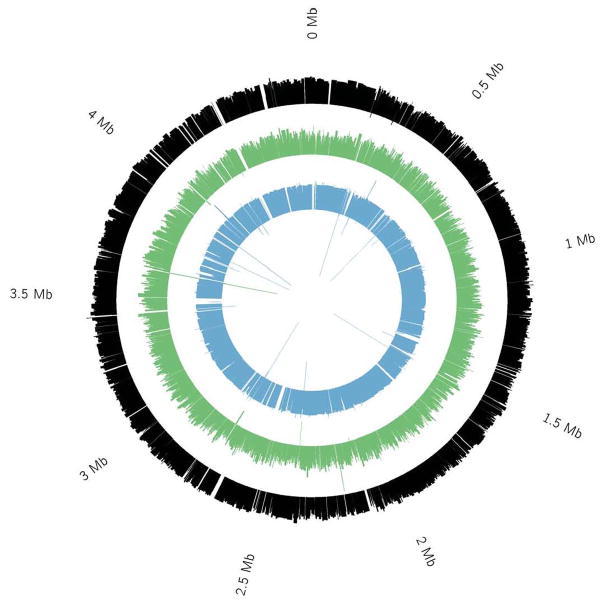
As a proof of concept, we used MIDAS to sequence three single MG1655 E. coli cells, generating 2 – 8 million paired-end Illumina MiSeq sequencing reads of 100 bp in length for each cell, which is equivalent to a genomic coverage of 87–364x. We first mapped the reads to the reference E. coli genome and recovered 98–99% of the genome at >1x coverage. Even when reads were downsampled such that genomic sequencing coverage was much lower (10x), we still recovered a high percentage of the genome (90%) (Supplementary Fig. 5). We then assembled the genome de novo using SPAdes11. We assembled 88–94% of the E. coli genome (Fig. 2), with an N50 contig size of 2,654 – 27,882 bp and a max contig length of 18,465 – 132,037 bp. More than 80% of the assembled bases were mapped to E. coli, with the remainder resulting from common MDA contaminants such as Delftia and Acidovorax (Supplementary Fig. 6, Supplementary Table 2). Despite the higher initial template concentration in the MIDAS libraries, chimerism was present at a comparable level to that previously reported for Illumina sequencing libraries constructed from conventional in-tube MDA reactions, with 1 chimeric junction per ~5 kb2 (Supplementary Table 3). We annotated the genome using the RAST and KAAS annotation servers. Over 96% of E. coli genes were either partially or fully covered in the assembly. Major biosynthetic pathways, including glycolysis and the citric acid cycle, were also present. Furthermore, pathways for amino acid synthesis and tRNA development were covered. MIDAS was thus able to assemble an extremely large portion of the E. coli genome from a single cell with comparatively minimal sequencing.
Figure 2.
Depth of coverage of assembled contigs aligned to the reference E. coli genome. Three single E. coli cells were analyzed using MIDAS. Between 88% and 94% of the genome was assembled from 2–8M paired-end 100bp reads. Each colored circle is a histogram of the log2 of average depth of coverage across each assembled contig for one cell. Gaps are represented by blank whitespace in between colored contigs
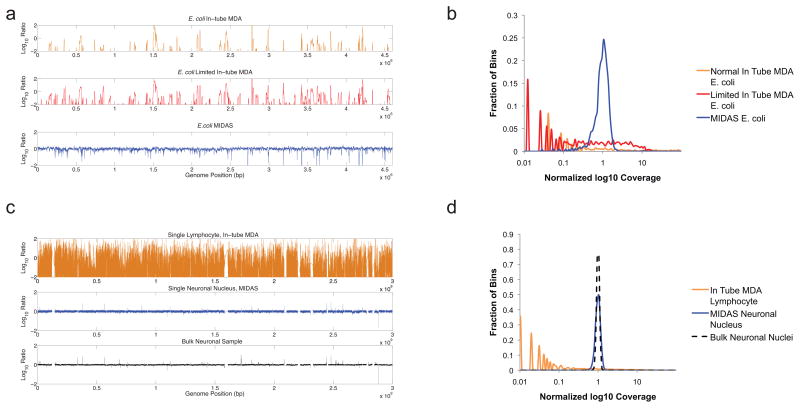
As a control, we also amplified and sequenced one E. coli cell using the conventional in-tube MDA method1, and controlled the reaction time to limit the amplification yield to the nanogram level. A fraction of the control amplicon was further amplified in a second reaction to the microgram level. The two control amplicons were converted into sequencing libraries using the conventional shearing and ligation method. We found that limiting the amplification yield reduced amplification bias, even for intube amplification. However, MIDAS had a markedly reduced level of amplification bias when compared with either control reaction (Fig. 3a,b). MIDAS was also able to recover a much larger fraction of the genome than the conventional MDA-based method. In fact, when compared with the most complete previously published single E. coli genome data set7, MIDAS recovered 50% more of the E. coli genome with 3 to 13-fold less sequencing data (~90–400x vs. ~1,200x). This result demonstrates that MIDAS provides a much more efficient way to assemble whole bacterial genomes from single cells without culture.
Figure 3.
Genomic coverage of single bacterial (a,b) and mammalian (c,d) cells amplified by MDA in a tube and by MIDAS. The observed multi-peak profile for the MDA reactions implies that certain regions may have been amplified with exponentially greater bias compared to the majority of the genome. (a) Comparison of single E. coli cells amplified in a PCR tube for 10 hours (top), 2 hours (middle) and in a microwell (MIDAS) for 10 hours (bottom). Genomic positions were consolidated into 1 kb bins (x-axis), and were plotted against the log10 ratio (y-axis) of genomic coverage (normalized to the mean). (b) Distribution of coverage of amplified single bacterial cells. The x-axis shows the log10 ratio of genomic coverage normalized to the mean. (c) Comparison of single human cells amplified using traditional MDA in a PCR tube for 10 hours (top) or in a microwell (MIDAS) for 10 hours (middle) to a pool of unamplified human cells (bottom). Genomic positions were consolidated into variable bins of approximately 60 kb in size previously determined to contain a similar read count28, and were plotted against the log10 ratio (y-axis) of genomic coverage (normalized to the mean). (d) Distribution of coverage of amplified single mammalian cells. The x-axis shows the log10 ratio of genomic coverage normalized to the mean.
Identification of copy number variants in single neurons
We next applied MIDAS to the characterization of copy number variation in single mammalian cells. The higher cognitive function of the human brain is supported by a complex network of neurons and glia. It has long been thought that all cells in a human brain share the same genome. Recent evidence suggests that individual neurons could have non-identical genomes owing to aneuploidy21–24, active retrotransposons25, 26 and other DNA content variation27. However, the presence of somatic genetic variation in individual neurons has not been conclusively demonstrated at the single-genome scale.
To demonstrate the viability of MIDAS as a tool for investigating copy number variation in single primary human neurons, we prepared nuclei from one post-mortem brain sample from a healthy female donor and a second post-mortem brain sample from a female individual with Down Syndrome. We purified cortical neuronal nuclei by flow sorting based on neuron-specific NeuN antibody staining. We generated six sequencing libraries (two disease-free and four Down Syndrome) from individual nuclei using MIDAS, and analyzed the data using a method based on circular binary segmentation to call copy number variation (CNV)28 (Supplementary Table 4). Raw sequencing reads were divided into 49,891 genomic bins ~60 kb in size, each of which had been previously determined to contain a similar number of sequencing reads in a fully diploid cell28. Although clonal read counts arising from PCR duplication appeared relatively high, this is a consequence of the low-input Nextera library construction protocol; because the amplification is limited, the amount of initial molecules is smaller, leading to more duplicates. However, the reduction in bias compensated for the apparent decrease in usable read count. We similarly observed a marked reduction of amplification bias in the MIDAS libraries when compared to the conventional in-tube MDA-based method (Fig. 3c,d). However, both MIDAS and intube MDA had higher levels of sequencing bias and variability than data generated from unamplified genomic DNA from 4,000 mammalian cells, though the bias in MIDAS was only slightly higher. Using a larger bin size of ~240kb (which results in a lower-resolution analysis) allowed MIDAS to match the level of bias from unamplified genomic DNA (Supplementary Fig. 7).
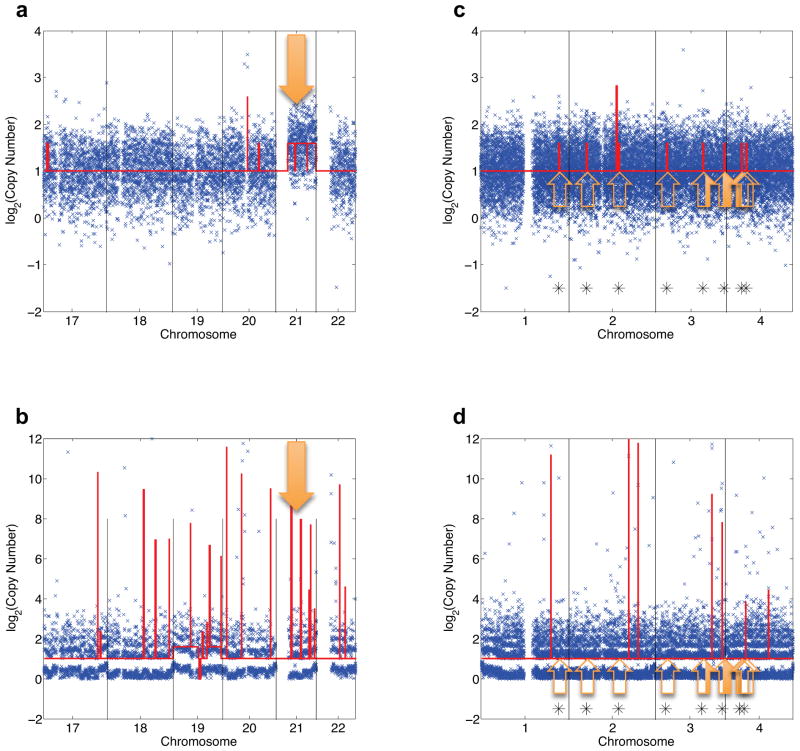
We next sought to characterize the sensitivity of detecting single copy-number changes. It was not possible to distinguish true copy number differences from random amplification bias for the conventional single-cell MDA data, even with aggressive binning into large genomic regions. However, the uniform genome coverage in the MIDAS libraries allowed clear detection of Trisomy 21 in each of the Down Syndrome nuclei (Fig. 4a, b). Rigorous validation of single-cell sequencing methods has been extremely challenging, primarily because any single cell might have genomic differences that are not detectable in the bulk cell population. Hence, there is no reference genome that single-cell data can be compared to. To determine the CNV detection limit of MIDAS, we computationally simulated sequencing data sets containing reference CNV events 1 or 2 Mb in size. We randomly selected 1 or 2 Mbps regions of either chromosome 21 (to simulate the gain of a single copy, the smallest possible copy number change) or chromosome 4 (as a negative control), and computationally transplanted these regions into 100 other random genomic locations (Supplementary Table 5). This computational approach, similar to a strategy previously used for assessing sequencing errors29, yielded data sets containing reference CNVs at known positions without affecting the inherent technical noise in the data. We identified 99/100 of 2 Mb T21 insertions and 80/100 of 1 Mb T21 insertions in the simulated data set from Down Syndrome Cell 1, indicating that MIDAS is able to call copy number events at the megabase-scale with high sensitivity (Fig. 4c, Supplementary Table 5). As expected, detection levels in the other data sets were similar for libraries with sufficient sequencing depth (80/100 for Down Syndrome Cell 2, 99/100 for Down Syndrome Cell 4), while libraries with insufficient sequencing depth could not be used for accurate small CNV calling (32/100 for Down Syndrome Cell 3). As expected, the insertion of diploid chromosome 4 regions did not generate any copy number calls. High-fidelity CNV calling (96%) at the 2 Mb level was retained even when 20% additional random technical noise was applied to the read count results (Supplementary Fig. 8). When the same simulation was performed with data from traditional in-tube MDA libraries, no T21 insertions were detected, indicating that at this level of sequencing depth, traditional MDA-based methods are unable to call small CNVs (Fig. 4d).
Figure 4.
Detection of copy number variants using MIDAS (a,c) and in-tube MDA (b,d). Genomic positions were consolidated into bins of approximately 60 kb in size which were previously determined to contain a similar read count28. Estimated copy numbers below were rounded to the nearest whole number. (a) Copy number variation in a Down Syndrome single cell analyzed with MIDAS. The x-axis shows genomic position, while the y-axis shows (on a log2 scale) the estimated copy number as a red line. The arrow indicates trisomy 21, which is clearly visible in this single cell. (b) Copy number variation in a Down Syndrome single cell analyzed with traditional in-tube MDA. The x-axis shows genomic position, while the y-axis shows (in a log2 scale) the estimated copy number as a red line. The arrow marks the expected region of Trisomy 21, which is not detectable in this data. (c) Copy number variation in a Down Syndrome single cell with Trisomy 21 “spike-ins.” The x-axis shows genomic position, while the y-axis shows (in a log2 scale) the estimated copy number as a red line. At each arrow, prior to CNV calling, data from a randomly determined 2 Mb section of Trisomy chromosome 21 was computationally inserted into the genome, simulating a small gain of single copy event. At each location, a copy number variant was called, showing that MIDAS can detect 2 Mb copy number variation accurately. (d) Copy number variation in a Down Syndrome single cell with Trisomy 21 “spike-ins.” The x-axis shows genomic position, while the y-axis shows (on a log2 scale) the estimated copy number as a red line. At each arrow, prior to CNV calling, data from a randomly determined 2 Mb section of Trisomy chromosome 21 was computationally inserted into the genome, simulating a small gain of single copy event.
We next performed CNV calling on each individual neuron using the parameters calibrated by the T21 transplantation simulation. MIDAS called 9–18 copy number events in each neuron (Supplementary Table 6). Only 8/60 called CNV events were larger than 2 Mb, and only 13/60 were larger than 1 Mb. It remained unclear whether the remaining events represented true copy number changes or whether they were false positives owing to the small size of most of the calls. It was also unclear which CNV calls represented somatic copy number variation and which represented germline CNV calls that might have been missed in one sample. To address these issues and further probe the ability of MIDAS to identify germline and de novo CNV events, we performed library construction and sequencing on unamplified genomic DNA from two pools of ~4,000 neuronal nuclei from the healthy donor, and compared the results to those obtained from the same donor’s single neuronal nuclei (Supplementary Table 7). We identified 22 CNV events in the unamplified libraries, of which only two were not shared between the two pools; these are likely false positive or false negative CNV calls in one sample. However, no CNV events identified in the pools were larger than 1 Mb. This finding is not surprising, as germline CNV events with size greater than 1 Mb do not commonly occur30. Although MIDAS does not have sufficient specificity when calling CNVs smaller than 1 Mb, we investigated how many small germline CNVs could be identified in the single cell libraries, and found that 75% were detected. Overall, based on the T21 computational transplantation results, it appears that the five individual human neurons (excluding Down Syndrome Cell 3 due to insufficient sequencing depth) contain an average of one region each with a somatic gain of one copy at the megabase scale, and that several smaller CNV events might also be present.
DISCUSSION
Owing to the extreme bias caused by whole-genome amplification from a single DNA molecule, genomic analysis of single cells has remained a challenging task. A large amount of sequencing resources is required to produce a draft-quality genome assembly or determine a low-resolution copy number variation profile owing to amplification bias and coverage dropout. MIDAS addresses this issue through the use of nanoliter-scale spatially confined volumes to generate nanogram-scale amplicons and the use of a low-input transposon-based library construction method. Compared to the conventional single-cell library construction and sequencing protocol, MIDAS provides a more-uniform, higher-coverage approach to analyze single cells from a heterogeneous population (Supplementary Table 8).
We applied MIDAS to single E. coli cells and resolved nearly the entire genome with relatively low sequencing depth. Additionally, using de novo assembly, >90 percent of the genome was assembled with far less sequencing effort than traditional MDA-based methods. These results suggest that applying MIDAS to an uncultivated organism would provide a draft quality assembly. Currently, a majority of unculturable bacteria are analyzed using metagenomics, as part of a mixed population rather than individually. Metagenomics has only recently allowed for the assembly of genomes from single cells, and doing so requires a sample with limited strain heterogeneity31. Through the use of MIDAS on heterogeneous environmental samples, novel single-cell organisms and genes can be easily discovered and characterized in a high-throughput manner, allowing a much higher-resolution and more complete analysis of single microbial cells.
We also applied MIDAS to the analysis of copy number variation in single human neuronal nuclei. With < 0.4x coverage, we used MIDAS to call single copy number changes of 1–2 million base pairs or larger in size. It has been shown recently that, in human adult brains, post-mitotic neurons in different brain regions exhibit various levels of DNA content variation (DCV)27. The exact genomic regions that associate with DNA content variation have been difficult to map to single neurons because of the amplification bias with existing MDA-based methods. CNVs in single tumor cells have been successfully characterized with a PCR-based whole-genome amplification method8. However, tumor cells tend to be highly aneuploid and exhibit copy number changes of larger magnitude, which are more easily detected. The applicability of a PCR-based strategy to other primary cell types with more subtle CNV events remains unclear. We have demonstrated that MIDAS greatly reduces the variability of single-cell analysis to a level such that a 1–2 Mb single-copy change is detectable, allowing characterization of much more subtle copy number variation. With additional improvements in sequencing methods, the use of MIDAS might enable the identification of even smaller CNVs, as currently 75% of smaller germline CNVs below the detection limit of MIDAS are still identifiable. Thirteen somatic gain of single copy events at the megabase level were identified in single neurons, and it appeared that several protease inhibitors, genes involved in vesicle formation, and genes involved in coagulation could be affected (Supplementary Table 7). A majority of gene copy changes occurred in one single cell, indicating that gene copy number might greatly vary across individual neurons. MIDAS can be used to simultaneously probe the individual genomes of many cells from patients with neurological diseases, and thus will allow identification of a range of structural genomic variants and eventually allow accurate determination of the influence of somatic CNVs on brain disorders in a high-throughput manner.
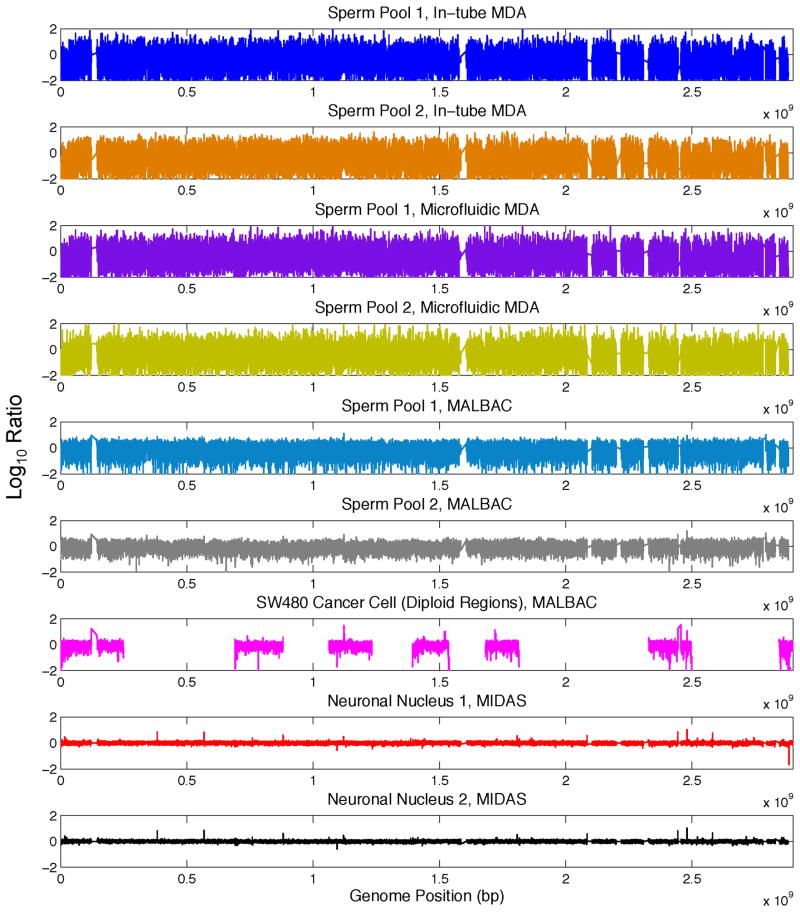
Recently, other single cell sequencing methods that reduce amplification bias and increase genomic coverage have been reported. One such method utilizes a microfluidic device to isolate single cells and perform whole genome amplification in a 60nL volume10. Another method, MALBAC, incorporates a novel enzymatic strategy to amplify single DNA molecules initially through quasi-linear amplification to a limited magnitude prior to exponential amplification and library construction18. MALBAC has been performed in microliter reactions in conventional reaction tubes. MIDAS represents an orthogonal strategy that adapts MDA to a microwell array. We compared data generated from single neurons amplified with MIDAS to previously published data from combined (and therefore diploid) pools of two single sperm cells amplified using standard in-tube MDA32, the microfluidic device10 and MALBAC18, 33. To ensure a fair comparison, we normalized sequencing depth to an equal amount for each method and processed the raw sequencing data for each sample using an identical computational pipeline. We also compared MIDAS to a single SW480 cancer cell amplified by MALBAC. In this case, to ensure a fair comparison to the primarily diploid cell analyzed using MIDAS, we limited our analysis to regions consistently identified as diploid in the cancer cell (parts of chromosomes 1, 4, 6, 8, 10 and 15)18. MIDAS compares favorably to each amplification method (Fig. 5, Supplementary Fig. 9), generating the lowest levels of bias across the genome.
Figure 5.
Comparison of MIDAS to previously published data for in-tube MDA35, microfluidic MDA10 and MALBAC36.for diploid regions of pools of two sperm cells and diploid regions of a single SW480 cancer cell processed using MALBAC34. Genomic positions were consolidated into variable bins of approximately 60 kb in size previously determined to contain a similar read count28, and were plotted against the log10 ratio (y-axis) of genomic coverage (normalized to the mean). For the cancer cell data, non-diploid regions have been masked out (white gaps between pink) to remove the bias generated by comparing a highly aneuploid cell to a primarily diploid cell.
Several aspects of MIDAS could be improved. First, the current efficiency of amplification is limited to 10%, owing to the use of a low cell–loading density to avoid having more than one cell per microwell. This efficiency could be improved 3 to 5 fold by increasing the cell loading density, imaging the microwell arrays containing fluorescently stained cells prior to amplification and excluding the wells with more than one cell from further analyses. Second, amplicon extraction by micromanipulation is currently performed manually at a speed of ~10 amplicons per hour. This number could be improved by at least one order of magnitude by implementing robotic automation. Third, the PDMS microwell arrays used for cell loading are highly customizable but require access to a microfabrication facility. Routine practice of MIDAS will depend on the commercial availability of hydrophilic microwell arrays. Finally, although each single cell is physically segregated into one microwell, the cells are not in total fluidic isolation. Thus, there may be the potential for cross-contamination between wells, and fluorescent imaging is required at least before and after MIDAS in order to ensure only single-cell amplicons are used.
MIDAS has the potential to provide researchers with a powerful tool for many other applications, including high-coverage end-to-end haplotyping of mammalian genomes or probing de novo CNV events at the single-cell level during the induction of pluripotency or stem cell differentiation34. MIDAS allows for efficient high-throughput sequencing of a variety of organisms. This technology should help propel single cell genomics, enhance our ability to identify diversity in multicellular organisms, and lead to the discovery of a multitude of new organisms in various environments.
METHODS
Microwell Array Fabrication
Microwell arrays were fabricated from polydimethylsiloxane (PDMS). Each array was 7 mm × 7 mm, with 2 rows of 8 arrays per slide and 255 microwells per array. The individual microwells were 400 μm in diameter and 100 μm deep (~12 nL volume), and were arranged in honeycomb patterns in order to minimize space in between the wells. To fabricate the arrays, first, an SU-8 mold was created using soft lithography at the Nano3 facility at UC San Diego. Next, a 10:1 ratio of polymer to curing agent mixture of PDMS was poured over the mold. Finally, the PDMS was degassed and cured for 3 hours at 65 °C.
Bacteria and Neuron Preparation
E. coli K12 MG1655 was cultured overnight, collected in log-phase, and washed 3x in PBS. After quantification, the solution was diluted to 10 cells/μL. Human neuronal nuclei were isolated as previously described27, 35 and fixed in ice-cold 70% ethanol. Nuclei were labeled with a monoclonal mouse antibody against NeuN (1:100 dilution) (Chemicon, Temecula, CA) and an AlexaFluor 488 goat anti-mouse IgG secondary antibody (1:500 dilution) (Life Technologies, San Diego, CA). Nuclei were counterstained with propidium iodide (50ug/ml) (Sigma, St. Louis, MO) in PBS solution containing 50 μg/ml RNase A (Sigma) and chick erythrocyte nuclei (Biosure, Grass Valley, CA). Nuclei in the G1/G0 cell cycle peak, determined by propidium iodide fluorescence, were electronically gated on a Becton Dickinson FACS-Aria II (BD Biosciences, San Jose, CA) and selectively collected based on NeuN+ immunoreactivity.
Cell Seeding, Lysis, and Multiple Displacement Amplification
All reagents not containing DNA or enzymes were first exposed to ultraviolet light for 10 minutes prior to use. The PDMS slides were treated with oxygen plasma to make them hydrophilic and ensure random cell seeding. The slides were then treated with 1% bovine serum albumin (BSA) (EMD Chemicals, Billerica, MA) in phosphate buffered saline (PBS) (Gibco, Grand Island, NY) for 30 minutes and washed 3x with PBS to prevent DNA from sticking to the PDMS. The slides were completely dried in a vacuum prior to cell seeding. Cells were diluted in 1x PBS to a concentration of 0.1 cells per well per array, and 3 μL of cell dilution was added to each array. This dilution ensures that approximately 99.5% of the wells have no more than one cell.
Initially, to verify that cell seeding adhered to the Poisson distribution, cells were stained with 1x SYBR green and viewed under a fluorescent microscope. Proper cell distribution was further confirmed with SEM imaging. For SEM imaging, chromium was sputtered onto the seeded cells for 6 seconds to increase conductivity. Note that the imaging of cell seeding was only used to confirm the theoretical Poisson distribution and not performed during actual amplification and sequencing experiments due to the potential introduction of contamination.
After seeding, cells were left to settle into the wells for 10 minutes. The seeded cells were then lysed either with 300 U ReadyLyse lysozyme at 100 U/μL (Epicentre, Madison, WI) and incubation at room temperature for 10 minutes, or with five 1 minute freeze/thaw cycles using a dry ice brick and room temperature in a laminar flow hood. After lysis, 4.5 μL of alkaline lysis (ALS) buffer (400 mM KOH, 100 mM DTT, 10 mM EDTA) was added to each array and incubated on ice for 10 minutes. Then, 4.5 μL of neutralizing (NS) buffer (666 mM Tris-HCl, 250 mM HCL) was added to each array. 11.2 μL of MDA master mix (1x buffer, 0.2x SYBR green I, 1 mM dNTP’s, 50 μM thiolated random hexamer primer, 8U phi29 polymerase, Epicentre, Madison, WI) was added and the arrays were then covered with mineral oil. The slides were then transferred to the microscope stage enclosed in a custom temperature controlled incubator set to 30 °C. Images were taken at 30-minute intervals for 10 hours using a 488 nm filter.
Image Analysis
Images were analyzed with a custom Matlab script to subtract background fluorescence. Because SYBR Green I was added to the MDA master mix, fluorescence under a 488 nm filter was expected to increase over time for positive amplifications. If a digital profile of fluorescent wells with increasing fluorescence over time was observed (approximately 10–20 wells per array), the array was kept. If no wells fluoresced, amplification failed and further experiments were stopped. Alternatively, if a majority of the wells fluoresced, the array was considered to have exogenous contamination from environmental DNA and subsequent analysis was similarly stopped. If 2 abutting wells fluoresced, neither was extracted due to the higher likelihood of more than one cell in each well existing (as in this case, seeding was potentially non-uniform). Finally, only wells with amplicons originating from a single point were extracted, ensuring that only single-cell derived amplicons were processed; thus, any potential cross-well contamination was prevented.
Amplicon Extraction
1 mm outer diameter glass pipettes (Sutter, Novato, CA) were pulled to ~30 um diameters, bent to a 45 degree angle under heat, coated with SigmaCote (Sigma, St. Louis, MO), and washed 3 times with dH20. Wells with positive amplification were identified using the custom Matlab script described above. A digital micromanipulation system (Sutter, Novato, CA) was used for amplicon extraction. The glass pipette was loaded into the micromanipulator and moved over the well of interest. The microscope filter was switched to bright field and the pipette was lowered into the well. Negative pressure was slowly applied, and the well contents were visualized proceeding into the pipette. The filter was then switched back to 488 nm to ensure the well no longer contained any fluorescent material. Amplicons were deposited in 1 μL dH20.
Amplicon Quantification
For quantification of microwell amplification, 0.5 μL of amplicon was amplified a second time using MDA in a 20 μL PCR tube reaction (1x buffer, 0.2x SYBR green I, 1 mM dNTP’s, 50 mM thiolated random hexamer primer, 8U phi29 polymerase). After purification using Ampure XP beads (Beckman Coulter, Brea, CA), the 2nd round amplicon was quantified using a Nanodrop spectrophotometer. The 2nd round amplicon was then diluted to 1 ng, 100 pg, 10 pg, 1 pg, and 100 fg to create an amplicon ladder. Subsequently, the remaining 0.5 μL of the 1st round amplicon was amplified using MDA along with the amplicon ladder in a quantitative PCR machine. The samples were allowed to amplify to completion, and the time required for each to reach 0.5x of the maximum fluorescence was extracted. The original amplicon concentration could then be interpolated. This 2nd round of MDA was only performed during amplicon quantification in order to determine approximately how much DNA was produced in each microwell. Amplicons that were sequenced were only subjected to the initial round of MDA, and thus did not have any secondary MDA or quantification performed.
Low-input library construction
1.5 μL of ALS buffer was added to the extracted amplicons to denature the DNA followed by a 3-minute incubation at room temperature. 1.5 μL of NS buffer was added on ice to neutralize the solution. 10 U of DNA Polymerase I (Invitrogen, Carlsbad, CA) was added to the denatured amplicons along with 250 nanograms of unmodified random hexamer primer, 1 mM dNTPs, 1x Ampligase buffer (Epicentre, Madison, Wi), and 1x NEB buffer 2 (NEB, Cambridge, MA). The solution was incubated at 37 °C for 1 hour, allowing second strand synthesis. 1 U of Ampligase was added to seal nicks and the reaction was incubated first at 37 °C for 10 minutes and then at 65 °C for 10 minutes. The reaction was cleaned using standard ethanol precipitation and eluted in 4 μL water.
Nextera transposase enzymes (Epicentre, Madison, WI) were diluted 100 fold in 1x TE buffer and glycerol. 10 μL transposase reactions were then conducted on the eluted amplicons after addition of 1 μL of the diluted enzymes and 1x tagment DNA buffer. The reactions were incubated for 5 minutes at 55 °C for mammalian cells and 1 minute at 55 °C for bacterial cells. 0.05 U of protease (Qiagen, Hilden, Germany) was added to each sample to inactivate the transposase enzymes; the protease reactions were incubated at 50 °C for 10 minutes followed by 65 °C for 20 minutes. 5 U Exo minus Klenow (Epicentre, Madison, WI) and 1 mM dNTP’s were added and incubated at 37 °C for 15 minutes followed by 65 °C for 20 minutes. Two stage quantitative PCR using 1x KAPA Robust 2G master mix (Kapa Biosystems, Woburn, MA), 10 μM Adapter 1, 10 μM barcoded Adapter 2 in the first stage, and 1x KAPA Robust 2G master mix, 10 μM Illumina primer 1, 10 μM Illumina primer 2, and 0.4x SYBR Green I in the second stage was performed and the reaction was stopped before amplification curves reached their plateaus. The reactions were then cleaned up using Ampure XP beads in a 1:1 ratio. A 6% PAGE gel verified successful tagmentation reactions.
Bulk Sample Library Construction
Genomic DNA was extracted from approximately 4,000 neuronal nuclei using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). The genomic DNA was incubated with 1 μL undiluted Nextera transposase enzymes and 1x tagment DNA buffer for 5 minutes at 55 °C. The reactions were cleaned with MinElute columns (Qiagen, Hilden, Germany) and eluted in 20 μL water. 5 U Exo minus Klenow (Epicentre, Madison, WI) and 1 mM dNTP’s were added and incubated at 37 °C for 15 minutes followed by 65 °C for 20 minutes. Two stage quantitative PCR using 1x KAPA Robust 2G master mix (Kapa Biosystems, Woburn, MA), 10 μM Adapter 1, 10 μM barcoded Adapter 2 in the first stage, and 1x KAPA Robust 2G master mix, 10 μM Illumina primer 1, 10 μM Illumina primer 2, and 0.4x SYBR Green I in the second stage was performed and the reaction was stopped before amplification curves reached their plateaus. The reactions were then cleaned up using Ampure XP beads in a 1:1 ratio. A 6% PAGE gel verified successful tagmentation reactions.
Mapping and De novo Assembly of Bacterial Genomes
Bacterial libraries were size selected into the 300–600 bp range and sequenced in an Illumina MiSeq using 100 bp paired end reads. E. coli data was both mapped to the reference genome and de novo assembled. For the mapping analysis_,_ libraries were mapped as single end reads to the reference E. coli K12 MG1655 genome using default Bowtie parameters with removal of any reads with multiple matches. Contamination was analyzed, and clonal reads were removed using SAMtools’ rmdup function. Chimeras were analyzed by flagging paired reads on the same strand or paired reads with a mismatched orientation. Chimeric junctions were defined as the number of chimeric reads divided by the total number of mapped bases. For the de novo assembly, paired end reads with a combined length less than 200 bp were first joined and treated as single end reads. All remaining paired end reads and newly generated single end reads were then quality trimmed. De novo assembly was performed using SPAdes11 v. 2.4.0. Corrected reads were assembled with kmer values of 21, 33, and 55. The assembled scaffolds were mapped to the NCBI nt database with BLAST, and the organism distribution was visualized using MEGAN36. Obvious contaminants (e.g., human) were removed from the assembly and the assembly was analyzed using QUAST37. The remaining contigs were annotated using RAST38 and KAAS39.
Identification of CNVs in MIDAS and MDA data
Mammalian single-cell libraries were sequenced in an Illumina Genome Analyzer IIx or Illumina HiSeq using 36 bp single end reads. The CNV algorithm previously published by Cold Spring Harbor Laboratories8 was used to call copy number variation on each single neuron, with modifications to successfully analyze non-cancer cells. Briefly, for each sample, reads were mapped to the genome using Bowtie. Clonal reads resulting from Polymerase Chain Reaction artifacts were removed using samtools, and the remaining unique reads were then assigned into 49,891 genomic bins of approximately 60 kb in size that were previously determined such that each would contain a similar number of reads after mapping28. Each bin’s read count was then expressed as a value relative to the average number of reads per bin in the sample, and then normalized by GC content of each bin using a weighted sum of least squares algorithm (LOWESS). Circular binary segmentation was then used to divide each chromosome’s bins into adjacent segments with similar means. Unlike the previously published algorithm, in which a histogram of bin counts was then plotted and the second peak chosen as representing a copy number of two, it was assumed, due to samples not being cancerous and thus being unlikely to contain significant amounts of aneuploidy, that the mean bin count in each sample would correspond to a copy number of two. Each segment’s normalized bin count was thus multiplied by two and rounded to the nearest integer to call copy number. MIDAS data clearly showed a CNV call designating Trisomy 21 in all Down Syndrome single cells, while the traditional MDA-based method was not able to call Trisomy 21.
Identification of Artificial CNVs in MDA and MIDAS data
In order to test the ability of the CNV algorithm described above to call small CNVs, artificial CNVs were computationally constructed. Prior to circular binary segmentation, in each Down Syndrome sample, one hundred random genomic regions across chromosomes 1–22 were chosen, each consisting of either 17 or 34 bins of approximately 60 kb in size. Each region was replaced with an equivalently sized region from chromosome 21 or chromosome 4 (Supplementary Table 5). The above algorithm was then run on each “spiked-in” sample, and the number of new CNV calls in each sample that matched each spike-in was tallied. For the chromosome 21 spike-ins, MIDAS was able to accurately call 98% of spiked-in CNVs at the 2 Mb level and 68% of spiked-in CNVs at the 1 Mb level, while the traditional MDA-based method was not able to call any spiked-in CNVs. As expected, spike-ins of chromosome 4 did not result in any additional CNV calls.
Supplementary Material
1
Acknowledgments
We thank C. Chen, H. Choi, and the UCSD Nano3 facility for initial help with microwell fabrication, F. Liang for initial technical assistance, P. Pevzner for advices on de novo genome assembly. This project was funded by NIH grants R01HG004876, R01GM097253, U01MH098977 and P50HG005550, and NSF grant OCE-1046368.
Footnotes
AUTHOR CONTRIBUTIONS
JG and KZ conceived and designed the experiments. JG, AR, and HIC performed the experiments. JG and YJC fabricated the microwell arrays. HLF performed sequencing. DB provided neuronal nuclei. JG, AG, and KZ analyzed data and wrote the manuscript with inputs from YHL and JC.
References
- 1.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigue S, et al. Whole genome amplification and de novo assembly of single bacterial cells. PLoS One. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 5.Pan X, et al. A procedure for highly specific, sensitive, and unbiased whole-genome amplification. Proc Natl Acad Sci U S A. 2008;105:15499–15504. doi: 10.1073/pnas.0808028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon HS, et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- 8.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chitsaz H, et al. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat Biotechnol. 2011;29:915–921. doi: 10.1038/nbt.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchison CA, 3rd, Smith HO, Pfannkoch C, Venter JC. Cell-free cloning using phi29 DNA polymerase. Proc Natl Acad Sci U S A. 2005;102:17332–17336. doi: 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcy Y, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue J, Shigemori Y, Mikawa T. Improvements of rolling circle amplification (RCA) efficiency and accuracy using Thermus thermophilus SSB mutant protein. Nucleic Acids Res. 2006;34:e69. doi: 10.1093/nar/gkl350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woyke T, et al. One bacterial cell, one complete genome. PLoS One. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzsimons MS, et al. Nearly finished genomes produced using gel microdroplet culturing reveal substantial intraspecies genomic diversity within the human microbiome. Genome Res. 2013 doi: 10.1101/gr.142208.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blainey PC, Quake SR. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 2011;39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adey A, Shendure J. Ultra-low-input, tagmentation-based whole-genome bisulfite sequencing. Genome Res. 2012;22:1139–1143. doi: 10.1101/gr.136242.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehen SK, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehen SK, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci U S A. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang AH, et al. Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J Neurosci. 2003;23:10454–10462. doi: 10.1523/JNEUROSCI.23-32-10454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yurov YB, et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS One. 2007;2:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- 26.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westra JW, et al. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol. 2010;518:3981–4000. doi: 10.1002/cne.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baslan T, et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 30.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albertsen M, et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 32.Kirkness EF, et al. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 2013;23:826–832. doi: 10.1101/gr.144600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hussein SM, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 35.Westra JW, et al. Aneuploid mosaicism in the developing and adult cerebellar cortex. J Comp Neurol. 2008;507:1944–1951. doi: 10.1002/cne.21648. [DOI] [PubMed] [Google Scholar]
- 36.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic acids research. 2007;35:W182–185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1