Simian Hemorrhagic Fever Virus Cell Entry Is Dependent on CD163 and Uses a Clathrin-Mediated Endocytosis-Like Pathway (original) (raw)
ABSTRACT
Simian hemorrhagic fever virus (SHFV) causes a severe and almost uniformly fatal viral hemorrhagic fever in Asian macaques but is thought to be nonpathogenic for humans. To date, the SHFV life cycle is almost completely uncharacterized on the molecular level. Here, we describe the first steps of the SHFV life cycle. Our experiments indicate that SHFV enters target cells by low-pH-dependent endocytosis. Dynamin inhibitors, chlorpromazine, methyl-β-cyclodextrin, chloroquine, and concanamycin A dramatically reduced SHFV entry efficiency, whereas the macropinocytosis inhibitors EIPA, blebbistatin, and wortmannin and the caveolin-mediated endocytosis inhibitors nystatin and filipin III had no effect. Furthermore, overexpression and knockout study and electron microscopy results indicate that SHFV entry occurs by a dynamin-dependent clathrin-mediated endocytosis-like pathway. Experiments utilizing latrunculin B, cytochalasin B, and cytochalasin D indicate that SHFV does not hijack the actin polymerization pathway. Treatment of target cells with proteases (proteinase K, papain, α-chymotrypsin, and trypsin) abrogated entry, indicating that the SHFV cell surface receptor is a protein. Phospholipases A2 and D had no effect on SHFV entry. Finally, treatment of cells with antibodies targeting CD163, a cell surface molecule identified as an entry factor for the SHFV-related porcine reproductive and respiratory syndrome virus, diminished SHFV replication, identifying CD163 as an important SHFV entry component.
IMPORTANCE Simian hemorrhagic fever virus (SHFV) causes highly lethal disease in Asian macaques resembling human illness caused by Ebola or Lassa virus. However, little is known about SHFV's ecology and molecular biology and the mechanism by which it causes disease. The results of this study shed light on how SHFV enters its target cells. Using electron microscopy and inhibitors for various cellular pathways, we demonstrate that SHFV invades cells by low-pH-dependent, actin-independent endocytosis, likely with the help of a cellular surface protein.
INTRODUCTION
Simian hemorrhagic fever virus (SHFV) is currently classified together with equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), and porcine reproductive and respiratory syndrome virus (PRRSV) in the genus Arterivirus, family Arteriviridae, in the order Nidovirales (1). The four arteriviruses are serologically distinct and cause remarkably different diseases in phylogenetically distant hosts. SHFV and SHFV-like viruses infect various African nonhuman primates without causing overt disease (2–5). In Asian macaques, however, SHFV causes a viral hemorrhagic fever that is nearly 100% lethal (6, 7).
Arterivirions are spherical to pleomorphic (40 to 55 nm in diameter) and enveloped and contain small surface protrusions (8). Like all arteriviruses, SHFV has a nonsegmented, linear, single-stranded RNA genome of positive polarity. The genome is polycistronic, capped at its 5′ end and polyadenylated at its 3′ end, and serves partially as an mRNA (9–12). Starting at the 5′ end, arterivirus genomes contain two plus-sense large open reading frames (ORFs 1a and 1b) that are directly translated into polyproteins pp1a and pp1ab. These polyproteins are autocatalytically cleaved into >12 nonstructural proteins that form the viral replicase complex that is also necessary for the synthesis of mRNA transcripts of the remaining, nested set of ORFs (reviewed in references 1 and 13). Similar to those of most nidoviruses, all SHFV mRNAs are 5′ and 3′ coterminal in sequence with the viral genome and are produced by discontinuous RNA transcription (12). These subgenomic mRNAs encode at least eight structural proteins that are essential for virion infectivity and appear to have functional analogs in particles of other arteriviruses (E, GP2 to -5, GP5a, M, and N) (reviewed in references 1 and 13). SHFV and SHFV-like viruses differ from EAV, LDV, and PRRSV by having four additional ORFs that may have emerged by duplication of existing ORFs coding for structural proteins (13, 14).
The molecular aspects of the SHFV life cycle have been understudied, but the more extensively characterized life cycles of the arteriviruses EAV and PRRSV are informative by analogy. SHFV N is an obvious homolog of the EAV and PRRSV nucleoprotein, which encapsidates the viral genome (12). The two major SHFV envelope proteins are the glycoprotein GP5 and the matrix protein M, which form heterodimers on the virion surface and contain the major neutralization epitopes (12, 15, 16). E is a myristoylated small integral envelope protein that may have ion channel properties and may facilitate virion uncoating (17). GP2, GP3, and GP4 are minor envelope glycoproteins that most likely form heterotrimers (18–20). E seems to be essential for insertion of this heterotrimer into the virion envelope (21). The functions of the recently discovered GP5a (20) and of the expression products of the additional ORFs found in SHFV and SHFV-like viruses remain to be determined.
GP2, GP3, GP4, and GP5 are suspected to engage the respective host cell surface receptor of each arterivirus. Because of the lack of structural similarities of these proteins to known class I to III fusion proteins (reviewed in reference 22), prediction of their exact functions is difficult. Host cell surface receptors have not been identified for any arterivirus, with the possible exception of PRRSV. Two cell surface factors, the macrophage-restricted sialoadhesin CD169/Sn/Siglec-1 and the more generally distributed CD163, have been implicated as possible receptors for PRRSV (23, 24), and PRRSV GP2 and GP4 interact with CD163 (25).
Only a few studies have attempted to elucidate the mechanism(s) by which arteriviruses gain entry into their host cells. In the cases of EAV and PRRSV, clathrin-mediated endocytosis (CME) seems to be the predominant route of cell entry (26–29). Here, we characterize the cell entry pathway of SHFV in the only SHFV-susceptible immortalized cell type, MA-104. Using transmission electron microscopy, chemical inhibitors, and antibody inhibition, we demonstrate that SHFV enters MA-104 cells via CD163-dependent low-pH endocytosis.
MATERIALS AND METHODS
Viruses and cells.
The SHFV prototype isolate LVR42-0/M6941 and PRRSV strain ISU-P were obtained from the American Type Culture Collection (ATCC) in Manassas, VA (VR-533 and VR-2402, respectively). SHFV and PRRSV stocks were prepared using the ATCC-recommended embryonic grivet monkey kidney cell line MA-104 (ATCC number CCL-2378), the only immortalized cell type known to support SHFV replication. All other cell lines reported to support SHFV replication, such as MARC-145, B-SC-1, and CL2621, are direct subclones of MA-104 cells. MA-104 cells and the subclone MARC-145 (provided by Kay Faaberg, U.S. Department of Agriculture, National Animal Disease Center, Ames, IA) were maintained in ATCC-recommended medium, Eagle's minimal essential medium (EMEM) (Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (SAFC Biosciences, Lenexa, KS), at 37°C in a humidified 5% CO2 atmosphere. The virus cultures obtained from the ATCC were designated passage 1 (p1). Confluent MA-104 cells were exposed to p1 viruses until the virus-induced cytopathic effect (CPE) affected 70 to 80% of the culture. The cells were lysed by two cycles of freeze-thawing, and the medium was clarified of debris by centrifugation (crude p2). Confluent MA-104 cells were exposed to p2 viruses at a multiplicity of infection (MOI) of 0.01 and observed until the CPE reached 70 to 80%. The cell supernatant was clarified of debris by centrifugation, and the virus was pelleted by ultracentrifugation (p3). The p3 virus and MA-104 cells were used for all the experiments described.
Recombinant vaccinia virus VV.NP-S-EGFP (VACV-eGFP) was kindly provided by Jonathan W. Yewdell (National Institutes of Health, Bethesda, MD). VACV-eGFP contains a chimeric gene encoding the influenza A virus nucleoprotein, the ovalbumin SIINFEKL peptide, and enhanced green fluorescent protein (eGFP) regulated by the P7.5 early-late promoter. VACV-eGFP stocks were prepared in BS-C-1 cells (ATCC number CCL-26) at an MOI of 0.01, as previously described (30).
Rift Valley fever virus strain ZH-548 MP12 (RVFV) stocks were prepared in Vero E6 cells (ATCC number CRL-1587) at an MOI of 0.01, as previously described (31).
Middle East respiratory syndrome coronavirus isolate HCoV-EMC/2012 (MERS-CoV) was kindly provided by the Erasmus Medical Center, Rotterdam, The Netherlands, and propagated on Vero E6 cells at an MOI of 0.01, as previously described (32).
SHFV infections and control virus infections.
Inoculation of MA-104 cells with SHFV (or the control virus MERS-CoV, RVFV, or VACV-eGFP) was performed uniformly across assays unless otherwise specified. First, the cell media were removed, and the cells were washed once with EMEM without FBS. The cells were then exposed to SHFV particles at the indicated MOIs and temperatures in a humidified 5% CO2 atmosphere for 1 h under gentle rocking every 15 min. The viral inocula were then removed, and the cells were washed once with EMEM without FBS and then incubated at 37°C in a humidified 5% CO2 atmosphere for the indicated times in EMEM (2% FBS).
SHFV quantification.
SHFV particle titers were quantified by plaque assay. Briefly, confluent layers of MA-104 cells in 6-well plates were exposed 24 h after seeding to serial dilutions of SHFV and incubated at 37°C for 1 h under gentle rocking every 15 min. The inocula were removed, and a 1% agarose overlay (ThermoFisher Scientific, Waltham, MA) was added. The infected cells were then incubated at 37°C for 48 h and stained with 5% neutral red (Sigma-Aldrich, St. Louis, MO). After incubating the cells at 37°C overnight, plaques were enumerated manually.
VACV-eGFP quantification.
Cells were infected with VACV-eGFP at an MOI of 1. The cells were washed two times with phosphate-buffered saline (PBS) and then grown in EMEM (2% FBS). At 6 h postinoculation, the cells were detached using trypsin and fixed with 4% paraformaldehyde (PFA) (Electron Microscopy Sciences, Hatfield, PA). The percentage of eGFP-positive cells (the percentage of cells infected) was measured by flow cytometry (BD LSR Fortessa; BD, San Jose, CA). Data analysis was performed using FlowJo software (Tree Star, Inc., San Carlos, CA).
MERS-CoV quantification.
Confluent monolayers of Vero E6 cells in 6-well plates were exposed to serial dilutions of MERS-CoV and incubated at 37°C for 1 h under gentle rocking every 15 min. The inocula were removed, and a 0.8% tragacanth overlay (Sigma-Aldrich) was added. The infected cells were incubated at 37°C for 72 h. The tragacanth overlay was removed and stained with 2% crystal violet (Sigma-Aldrich) in 10% neutral buffered formalin (Fisher Scientific, Kalamazoo, MI). Plaques were enumerated manually.
RVFV quantification.
At 16 h postinfection, RVFV-infected MA-104 cells were fixed with 4% PFA and then stained with anti-RVFV Gn 4D4 monoclonal antibody (MAb) obtained from the United States Army Medical Research Institute of Infectious Diseases (USAMRIID), Fort Detrick, Frederick, MD, at a 1:1,000 dilution for 2 h at 37°C, followed by staining with Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (Life Technologies, Grand Island, NY). The Operetta high-content imaging system (PerkinElmer, Shelton, CT) was used for quantitative imaging analysis with Harmony 3.1 analysis software (PerkinElmer).
Transmission electron microscopy.
Confluent MA-104 cells were exposed to SHFV particles (as described above) at an MOI of 50 for 1 h at 4°C to permit virion adsorption to cell surfaces. The inocula were removed, and the cells were incubated at 37°C for 0, 5, 10, 20, or 40 min. The media were removed, and electron microscopy grade fixative, 2.5% glutaraldehyde (Electron Microscopy Sciences) in Millonig's sodium phosphate buffer (Tousimis Research, Rockville, MD), was added directly to the flasks. The cells were scraped off 10 min later with a cell scraper, collected into 15-ml tubes, and immediately centrifuged at 500 × g for 20 min. The fixed cells were kept for 24 h at 4°C to complete fixation. The preserved cells were postfixed in 1% osmium tetroxide (Electron Microscopy Sciences), stained en bloc with 2% uranyl acetate, dehydrated in a graded ethanol series, and infiltrated and embedded in Spurr plastic resin (Electron Microscopy Sciences). The embedded blocks were sectioned using a Leica EM UC7 ultramicrotome (Leica Microsystems, Buffalo Grove, IL), and 60- to 80-nm ultrathin sections were collected, mounted on 200-mesh copper grids, and contrasted with lead citrate. The grids were then examined and imaged using an FEI G2 Tecnai transmission electron microscope (FEI, Hillsboro, OR) operating at 80 kV.
Inhibitor studies.
Ammonium chloride (NH4Cl), blebbistatin, chloroquine, CA-074, chlorpromazine, α-chymotrypsin, concanamycin A, cytochalasin B, cytochalasin D, dynasore, E-64d, 5-(_N_-ethyl-_N_-isopropyl) amiloride (EIPA), filipin III, FYdmk, latrunculin B, methyl-β-cyclodextrin, monensin, nystatin, phenylarsine oxide, phospholipase A2, papain, phospholipase D, proteinase K, trypsin, and wortmannin were obtained from Sigma-Aldrich. Dyngo-4a was synthesized in house (Adam McCluskey).
Confluent cells were exposed to the indicated concentrations of inhibitors in medium for 30 min (ammonium chloride, blebbistatin, chloroquine, chlorpromazine, concanamycin A, cytochalasin B, cytochalasin D, dynasore, Dyngo-4a, EIPA, filipin III, latrunculin B, monensin, nystatin, phenylarsine oxide, and wortmannin), 1 h (α-chymotrypsin, methyl-β-cyclodextrin, papain, phospholipase A2, phospholipase D, proteinase K, and trypsin), or 4 h (CA-074, E-64d, and FYdmk). The media containing inhibitors were removed, and the cells were washed twice with EMEM (10% FBS) (methyl-β-cyclodextrin, phospholipase A2, phospholipase D, trypsin, proteinase K, papain, and α-chymotrypsin) to remove enzyme activity. Alternatively, virus was directly added to inhibitor-containing media (ammonium chloride, CA-074, chloroquine, concanamycin A, blebbistatin, chlorpromazine, cytochalasin B, cytochalasin D, dynasore, Dyngo-4a, E-64d, EIPA, filipin III, FYdmk, latrunculin B, monensin, nystatin, phenylarsine oxide, and wortmannin). The cells were then exposed to SHFV for 1 h at an MOI of 5, followed by plaque assay quantification 18 h later.
For time-of-addition experiments, MA-104 cells were placed on ice and exposed to SHFV at an MOI of 5 for 1 h to allow virion adsorption. Unbound virions were washed off, 50 μl of EMEM was added, and the plates were moved to 37°C. Zero, 5, 10, 15, 30, 45, 60, 75, 90, 105, or 120 min later, 50 μl of EMEM with 300 μM chloroquine (final concentration, 150 μM) or 30 μM Dyngo-4a (final concentration, 15 μM) were added to the plates, and incubation at 37°C was continued until a total time span of 3 h was reached. Then, the inhibitor mixture was removed, the cells were washed, EMEM (2% FBS) was added, and the cells were incubated for 18 h before virus production was measured in supernatants using the plaque assay.
The cytotoxicity of inhibitors was assessed using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Rockville, MD), according to the instructions of the manufacturer, on uninfected cells in parallel with infection assays.
Dynamin mutant transfection.
Plasmids encoding dynamin-green fluorescent protein (GFP) (pEGFP-Dynamin1-WT) and the dominant-negative mutant dynamin-K44A-GFP (pEGFP-Dynamin1-K44A) were kindly provided by Robert A. Davey (Texas Biomedical Research Institute, San Antonio, TX). MA-104 cells were transfected with plasmids using Lipofectamine 3000 (Life Technologies) according to the manufacturer's instructions. Forty-eight hours after transfection, the cells were detached with Accumax (Innovative Cell Technologies, San Diego, CA), and GFP-expressing cells were sorted using a BD FACS Aria II cell sorter (BD Biosciences, San Jose, CA) and reseeded. After recovery, the sorted GFP-expressing cells were infected with SHFV at an MOI of 1. The cell culture supernatants were harvested 20 h postinoculation, and virus titers were measured by plaque assay.
Clathrin heavy chain knockout experiments.
CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 technology was used to knock out clathrin heavy chain (CLTC-HC). CLTC-HC genes from members of different primate species were aligned to choose conserved regions. The DNA2.0 CRISPR gRNA Design Tool (DNA 2.0, Menlo Park, CA) was used to design guide RNAs (gRNAs). The gRNAs (gRNA-84, gRNA1 [CATGGCGCTGTCGGGGGTTA] and gRNA2 [GATTCTGCCAATTCGTTTTC]; gRNA-85, gRNA1 [AATCTGGGCCATGGCGCTGT] and gRNA2 [TCGTTTTCAGGAGCATCTCC]; and gRNA-86, gRNA1 [AGATGCTCCTGAAAACGAAT] and gRNA2 [CGGCCGGGCCCGGGCTGGTG]) were synthesized and incorporated into suitable vectors (pD1411-AD: CBh-Cas9N-2A-GFP, Cas9-ElecD) by DNA2.0, resulting in plasmids pgRNA-CLTC-84 and pgRNA-CLTC-85 and three pgRNA-CLTC-86 plasmids. MA-104 cells were transfected with these plasmids using Lipofectamine 3000 and dissociated with Accumax 48 h later. The GFP-expressing cells were sorted using a BD FACS Aria II cell sorter, split into two sets, and reseeded. After recovery, the first set of cells was washed with PBS and lysed in cell lysis buffer (Cell Signaling, Danvers, MA) according to the manufacturer's instructions.
Protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. Equivalent amounts of total cellular lysates were resolved in 4% to 12% Bis-Tris gradient gels (Life Technologies) and then dry transferred to polyvinylidene difluoride (PVDF) membranes (Life Technologies) by using the iBlot gel transfer system (Life Technologies). After blocking in 5% nonfat milk powder in PBS with 0.1% Tween (Sigma-Aldrich), the membranes were incubated overnight with goat anti-clathrin HC antibody (1:500; SICGEN, Cantanhede, Portugal) or anti β-actin antibody (1:500; Abcam, Cambridge, MA), followed by incubation with appropriate horseradish peroxidase-conjugated secondary antibodies (Sigma-Aldrich). Signals were detected with a SuperSignal West Femto chemiluminescent substrate (Pierce), and images were acquired using a Syngene G:Box Chemiluminescence imaging system (Syngene, Frederick, MD). The second set of sorted GFP-expressing cells were infected with SHFV at an MOI of 1. The cell culture supernatants were harvested 20 h postinoculation, and virus titers were measured by plaque assay.
Actin staining.
MA-104 cells were treated with inhibitors at 37°C for 1 h and then washed twice with warm PBS (Gibco, Carlsbad, CA). The cells were then fixed with 4% PFA and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) in PBS. The cells were stained with Alexa 594-phalloidin (Life Technologies) for 20 min at room temperature and again washed twice with PBS, mounted, and viewed.
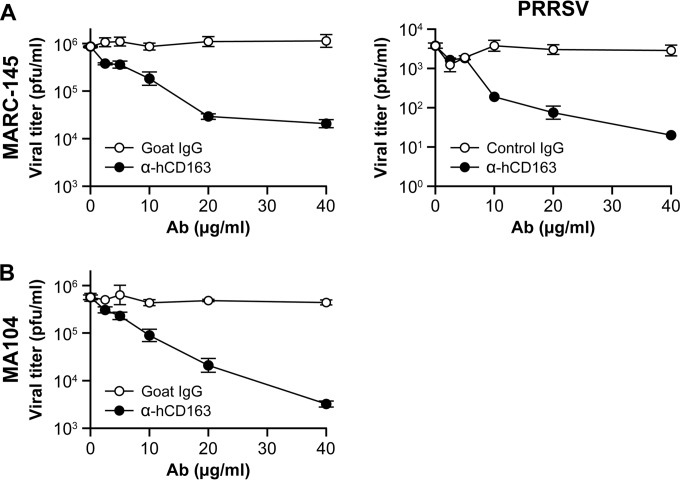
CD163 antibody inhibition assay.
MA-104 or MARC-145 cells were incubated with different concentrations (40, 20, 10, 5, 2.5, and 0 μg/ml) of goat anti-human CD163 antibody (R&D Systems, Minneapolis, MN) or control goat IgG at 37°C for 1 h. The treated cells were infected with SHFV or PRRSV at an MOI of 5 at 37°C for 1 h in the presence of antibodies. The virus-antibody inocula were then removed, the cells were washed, and EMEM (2% FBS) was added. The supernatants were harvested 24 h postinoculation, and viral titers were determined by plaque assay.
RESULTS
SHFV particles rapidly enter target cells.
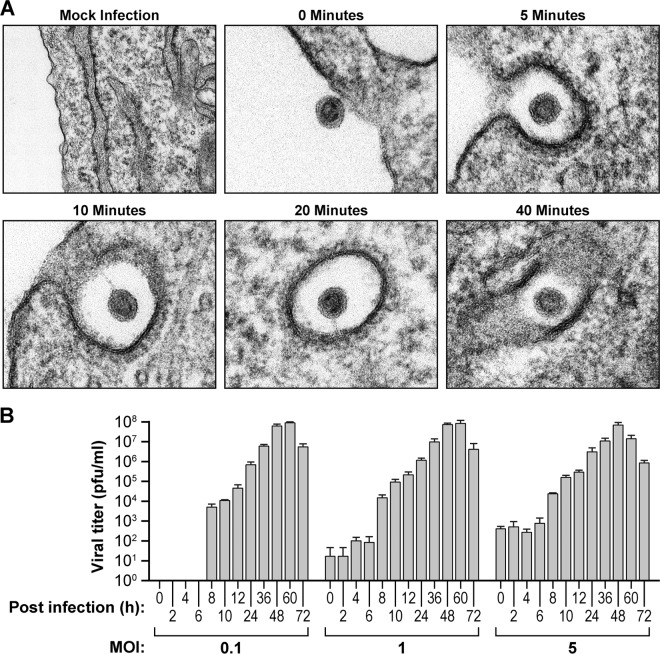
To determine how quickly SHFV particles enter target cells, we exposed MA-104 cells to virions at an MOI of 50 for 1 h at 4°C to allow particle cell surface adsorption. We then warmed the cells to 37°C for 0, 5, 10, 20, or 40 min and examined the cells through conventional thin-section ultrastructural electron microscopy. Our analysis revealed the formation of electron-dense cell membrane invaginations around viral particles that in appearance were reminiscent of clathrin-coated vesicles (Fig. 1A). These particles entered cells within the first 5 min after virion adsorption. The majority of virus particles were found to be completely engulfed by the target cells 20 min postinoculation, although a small number of particles were engulfed as late as 40 min. Next, we determined the time frame of the SHFV replication cycle. We incubated MA-104 cells with SHFV particles at an MOI of 0.1, 1, or 5 for 1 h at 37°C; removed the inocula; and incubated the cells for 0, 2, 4, 6, 8, 10, 12, 24, 36, 48, 60, or 72 h in fresh medium. Viral titers in cell supernatants (and throughout the study) were determined by plaque assay, as reporter gene-encoding cDNA clones of SHFV, virion-like particles pseudotyped with SHFV surface proteins, and SHFV-specific antibodies suitable for indirect fluorescence assays (IFA) are yet to be established. For all MOIs, viral titers increased more than 10- to 100-fold over baseline at 8 h, with viral production peaking at 48 to 60 h (Fig. 1B). These data indicate that SHFV completes its life cycle in as little as 8 h and that interference with SHFV entry mechanisms could be measured before this time point by plaque assay.
FIG 1.
SHFV particles rapidly enter target cells. (A) Time-dependent transmission electron micrographs of SHFV particle entry into MA-104 cells. Shown are representative images of two independent experiments. (B) Growth kinetics of SHFV at various MOIs in MA-104 cells as determined by plaque assay. The error bars indicate the standard deviations of triplicate samples from one of two independent experiments.
SHFV particles enter target cells in a low-pH-dependent manner.
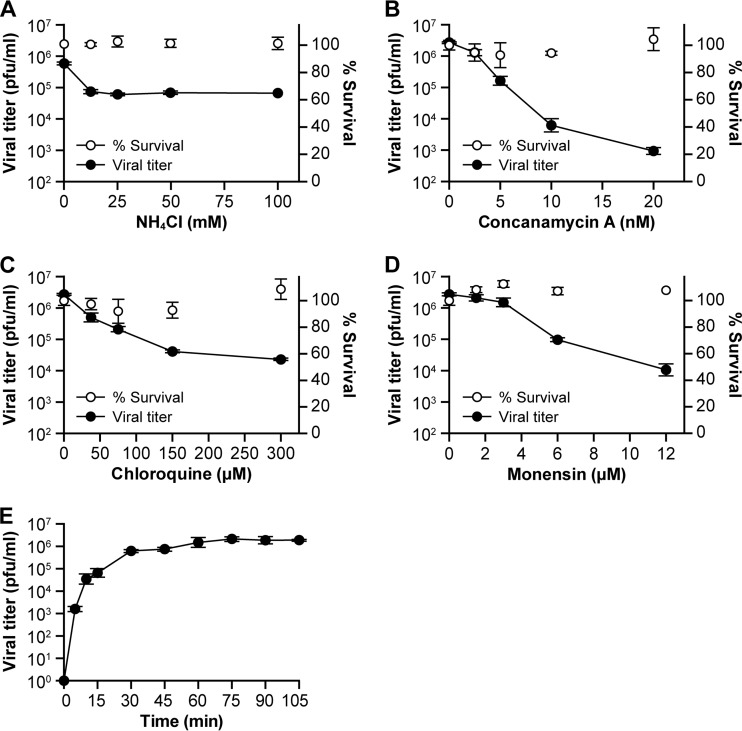
To determine the effect of pH on SHFV infectivity, we exposed MA-104 cells to increasing concentrations of ammonium chloride, chloroquine, concanamycin A, or monensin for 30 min. The cells were exposed to SHFV at an MOI of 5 for 1 h and then incubated at 37°C for 20 h for titer measurement. Ammonium chloride and chloroquine, both lysosomotropic weak bases, and monensin, a carbocyclic polyether Na+ ionophore that inhibits endosomal acidification by acting as an Na+/H+ antiporter (33), reduced SHFV particle production by roughly 10- to 100-fold at concentrations of 12.5 mM, 150 μM, and 12 μM, respectively. Concanamycin A, a specific inhibitor of vacuolar-type H+-ATPases and thus a strong inhibitor of endosomal acidification (34), reduced SHFV particle production by roughly 5,000-fold at a concentration of only 20 nM (Fig. 2A to D). Together, these observations indicate that SHFV uses a pH-dependent and, therefore, most likely an endosomal cell entry pathway.
FIG 2.
SHFV particles enter target cells in a low-pH-dependent manner. (A to D) Effects of pretreatment of MA-104 cells with increasing concentrations of inhibitors affecting pH on SHFV particle yield, as determined by plaque assay. (E) Time-of-addition experiment using chloroquine at a fixed concentration of 150 μM. The error bars indicate the standard deviations of triplicate samples from one of two independent experiments.
Next, we performed time-of-addition experiments with chloroquine at a concentration of 150 μM. Chloroquine's inhibitory effect on SHFV particle production was maximal when the drug was added to target cells directly after virus adsorption and progressively decreased until it was lost ∼30 min later (Fig. 2E). These results further substantiate the rapidity of SHFV particle cell entry observed by electron microscopy (Fig. 1A) and confirm that virus entry is pH dependent.
SHFV particles enter cells using a clathrin-mediated endocytosis-like pathway.
Several pH-dependent endocytic pathways are used by viruses, including dynamin-mediated (usually clathrin-mediated) endocytosis, clathrin-independent endocytosis, macropinocytosis, and caveola-mediated endocytosis (35).
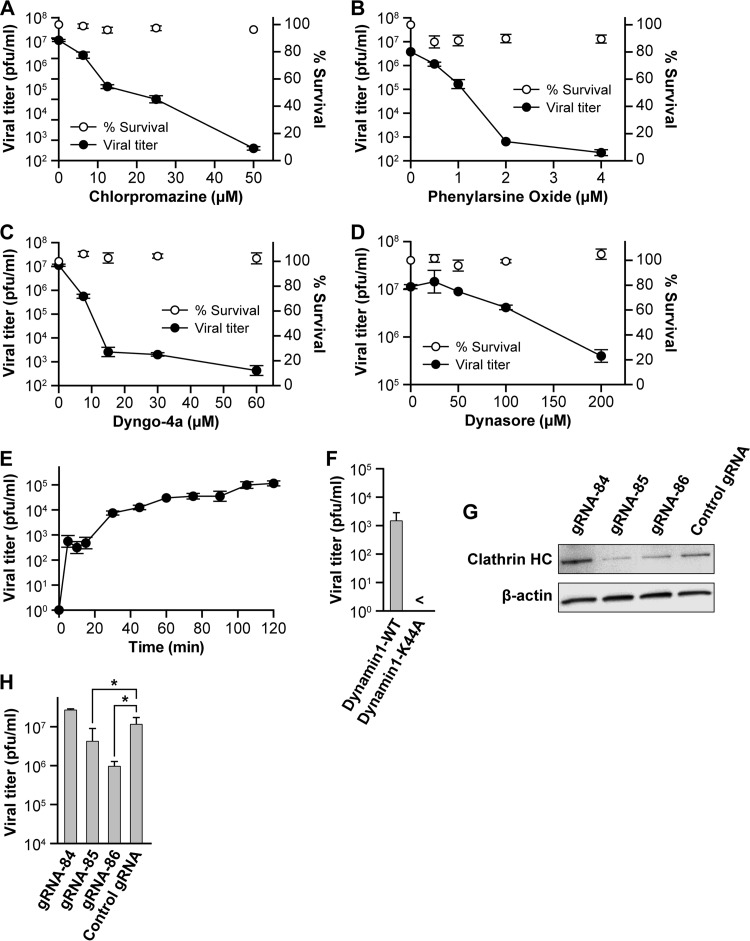
PRRSV and EAV both enter cells via CME, and our transmission electron microscopy data suggest that SHFV may utilize a similar mechanism (Fig. 1A). Therefore, prior to SHFV infection, we treated MA-104 cells with increasing concentrations of chemicals known to inhibit CME. Chlorpromazine, suggested to abolish the formation of clathrin-coated endocytic vesicles by interfering with the interaction between the adapter protein AP-2 and the clathrin-coated-pit lattice (36), almost completely inhibited SHFV particle production at a concentration of 50 μM (Fig. 3A). A similar effect was observed with phenylarsine oxide, which inhibits CME at concentrations of 1 to 20 μM by interfering with the production of phosphatidylinositol 4-phosphate, the precursor of phosphatidylinositol 4,5-bisphosphate, needed for CME (37). In this study, phenylarsine oxide inhibited CME at concentrations as low as 1 to 2 μM (Fig. 3B).
FIG 3.
SHFV particles enter cells using a clathrin-mediated endocytosis-like pathway. (A to D) Effects of pretreatment of MA-104 cells with increasing concentrations of inhibitors of clathrin-mediated endocytosis on SHFV particle yield as determined by plaque assay. (E) Time-of-addition experiment testing the effects of Dyngo-4a at the same concentration but at different points after cell exposure to SHFV particles. (F) Effect of overexpression of wild-type dynamin 1 or a dominant-negative mutant thereof in MA-104 cells on SHFV progeny production. (G) Evaluation of clathrin HC expression in MA-104 cells treated with clathrin HC-specific gRNAs or control gRNA by Western blotting. (H) Effects of gRNA treatment of MA-104 cells on SHFV progeny production. The error bars indicate the standard deviations of triplicate samples of one of two independent experiments. <, measurement below the threshold of detection (20 PFU/ml); *, P < 0.05 (Student's t test).
Dynamin acts as a mechanochemical enzyme driving membrane fission and as a regulatory GTPase in CME and in clathrin-independent dynamin-dependent endocytosis (38, 39). Dynasore and Dyngo-4a are both inhibitors of dynamin, and these inhibitors reduced SHFV particle production (Fig. 3C and D). Dynasore reduced SHFV titers by more than 10-fold at high (200 μM) concentrations, whereas at a concentration of 15 μM, Dyngo-4a reduced SHFV titers by almost 10,000-fold. Next, we performed Dyngo-4a time-of-addition experiments to evaluate whether Dyngo-4a acts at an early time point of SHFV infection. We treated MA-104 cells with the drug at different time points after virus adsorption, followed by SHFV progeny quantification by plaque assays. Not surprisingly, a dramatic inhibitory effect of Dyngo-4a on SHFV production was observed when the drug was added immediately after virus adsorption. However, the drug lost most of its inhibitory effect within 30 min (Fig. 3E). This observation indicated that Dyngo-4a acts within the first 30 min following virus adsorption and, therefore, during the viral entry stage of the SHFV life cycle.
To further substantiate the possible involvement of dynamin in SHFV cell entry, we transfected MA-104 cells with expression plasmids encoding wild-type dynamin-GFP (pEGFP-Dynamin1-WT) or the dominant-negative mutant dynamin-K44A-GFP (pEGFP-Dynamin1-K44A). We sorted the cells 48 h later for GFP expression, infected the sorted cells with SHFV at an MOI of 1, and quantified SHFV production in cell culture supernatants 20 h later by plaque assay. Expression of the dominant-negative mutant Dynamin1-K44A completely abrogated SHFV progeny production (Fig. 3F). We also hypothesized that clathrin heavy chain is a crucial component of the SHFV cell entry process. We employed CRISPR/Cas9 technology, which is based on the expression of the bacterial RNA-guided DNA endonuclease Cas9, and a targeting single gRNA to direct cleavage of a target gene (40, 41) by following standard protocols (42). We transfected three gRNAs designed to target the clathrin heavy chain gene into MA-104 cells prior to infection with SHFV. Two gRNAs (gRNA-85 and gRNA-86) reduced clathrin heavy chain expression as judged by Western blotting (Fig. 3G). SHFV progeny production was impaired in cells pretreated with these RNAs but not in cells pretreated with gRNA-84 or control gRNA (Fig. 3H), suggesting that clathrin heavy chain plays a role in SHFV cell entry.
Together, the data presented in Fig. 3 suggest that SHFV enters target cells by dynamin-mediated endocytosis, most likely reflecting CME.
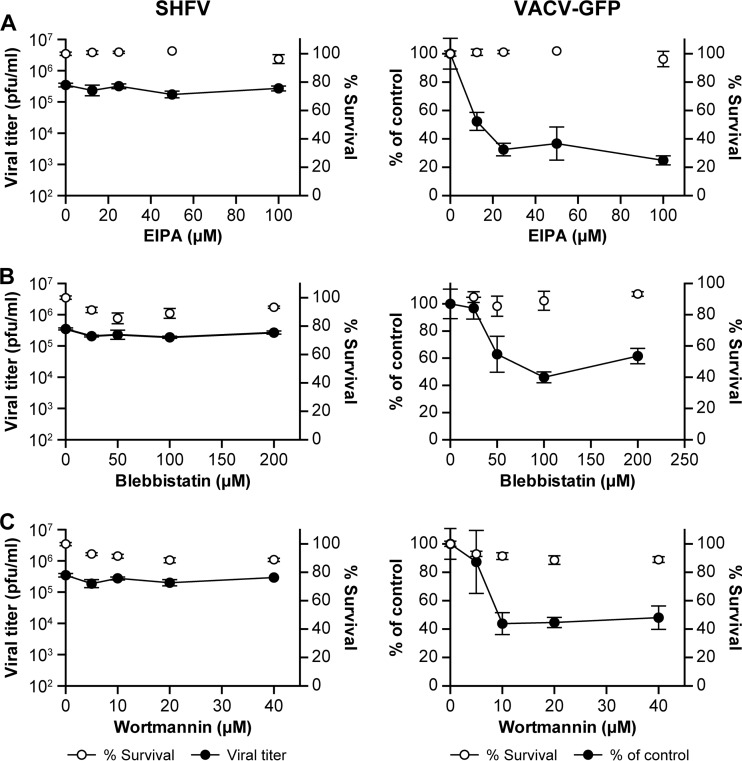
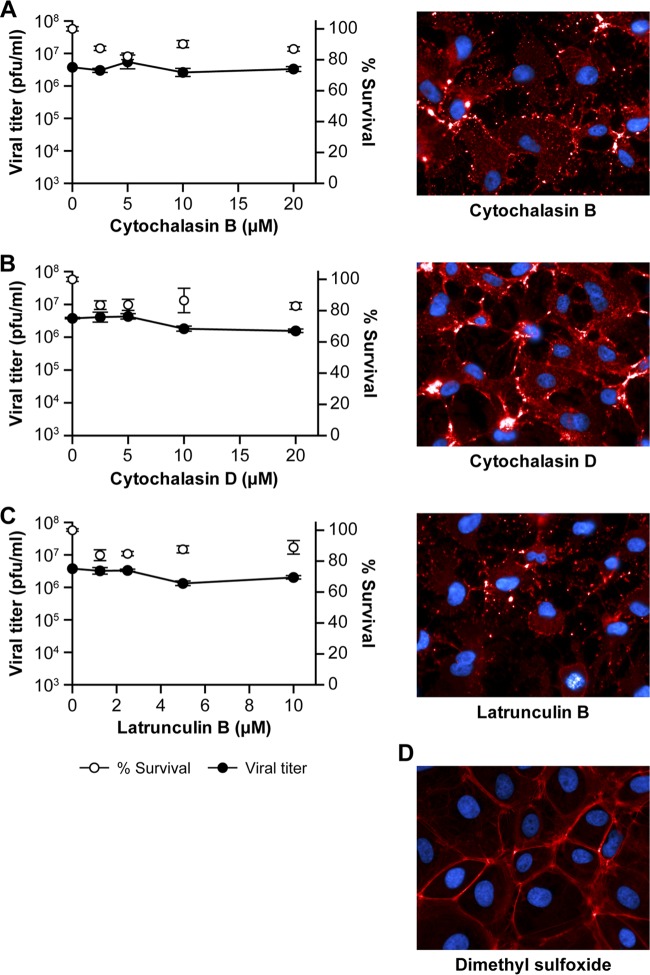
To examine the possibility that SHFV enters target cells via macropinocytosis, we treated MA-104 cells with increasing concentrations of compounds known to inhibit this type of endocytosis (i.e., EIPA, blebbistatin, and wortmannin) prior to the addition of virus. EIPA is an Na+/H+ antiport inhibitor, blebbistatin lowers the affinity of myosin II for actin and, therefore, inhibits macropinocytosis-characteristic plasma membrane bleb formation (43), and wortmannin is a covalent inhibitor of phosphatidylinositide 3-kinases. None of the three compounds had an effect on SHFV particle production, as indicated by plaque assay (Fig. 4A to C, left). However, the inhibitors had an effect on the percentage of VACV-eGFP-infected cells, as indicated by flow cytometry (Fig. 4A to C, right). Importantly, VACV-eGFP infection did not appear to be reduced in the presence of these inhibitors when measured by plaque assays (data not shown), demonstrating that plaque assays may not be sensitive enough to detect the effects of macropinocytosis inhibitors. As macropinocytosis is strongly dependent on actin, we treated MA-104 cells with increasing concentrations of cytochalasin B, cytochalasin D, or latrunculin B, potent inhibitors of actin polymerization. To verify that all three compounds inhibited actin polymerization, we stained uninfected, but compound-treated, MA-104 cells with Alexa 594-coupled phalloidin. The three compounds potently disrupted the cellular actin filament network but had little effect on SHFV particle production (Fig. 5A to C). Together, these data, and the absence of plasma membrane ruffling or blebbing in SHFV-infected MA-104 cells (Fig. 1A), suggest that macropinocytosis does not play a major role in SHFV target cell entry.
FIG 4.
SHFV particles do not enter cells by macropinocytosis. (A to C) Effects of pretreatment of MA-104 cells with increasing concentrations of inhibitors of macropinocytosis on the SHFV viral titer, as determined by plaque assay (left), or on the percentage of VACV-GFP-infected cells, as determined by high-content imaging (right). The error bars indicate the standard deviations of triplicate samples from one of two independent experiments.
FIG 5.
SHFV cell entry is independent of actin polymerization. (A to C) (Left) Effects of pretreatment of MA-104 cells with increasing concentrations of inhibitors of actin polymerization on SHFV particle yield, as determined by plaque assay. (Right) Immunofluorescence images of the MA-104 cells showing the disruption of actin networks by the inhibitors using Alexa 594-phalloidin staining. (D) Alexa 594-phalloidin staining of untreated MA-104 cells treated with dimethyl sulfoxide (DMSO). The error bars indicate the standard deviations of triplicate samples from one of two independent experiments.
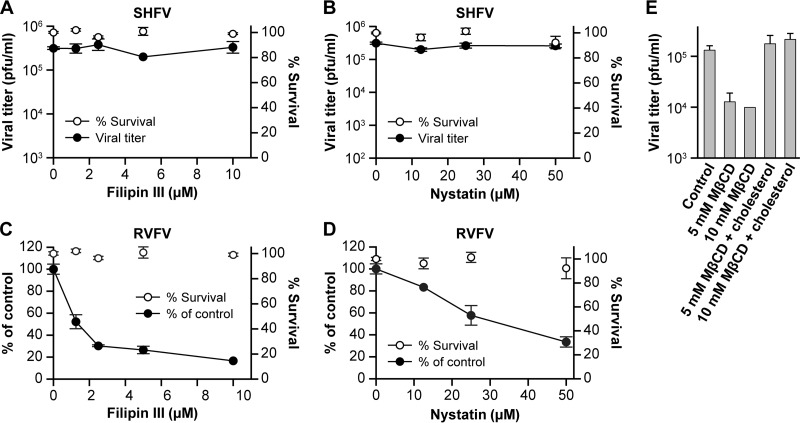
Next, we used chemicals that inhibit the caveolin pathway to test whether SHFV enters cells via this pathway. Specifically, we treated MA-104 cells with increasing concentrations of the membrane disruptor filipin III and the cholesterol sequestrator nystatin. While both inhibitors decreased the RVFV infection rate, as previously described (44), neither drug had any effect on SHFV particle production, as judged by plaque assay (Fig. 6A to D). The effect of the inhibitors on RVFV infection was too weak to be detected in plaque assays (data not shown), leaving room for the possibility that the inhibitors do have an effect on SHFV infection that we could not measure.
FIG 6.
SHFV particles do not enter cells by caveola-mediated endocytosis. (A and B) Effects of pretreatment of MA-104 cells with increasing concentrations of inhibitors of caveola-mediated endocytosis on SHFV particle yield, as determined by plaque assay. (C and D) Effects of pretreatment of MA-104 cells with increasing concentrations of the same inhibitors on RVFV infection, as determined by IFA. (E) Influence of cholesterol depletion on SHFV particle yield. Cells were treated with 5 or 10 mM methyl-β-cyclodextrin (MβCD) and infected with SHFV. The particle yield was determined by plaque assay. Alternatively, the cells were treated with MβCD, and exogenous soluble cholesterol was added to reverse the effect of MβCD. The error bars indicate the standard deviations of triplicate samples from one of two independent experiments.
Treatment of MA-104 cells with the cholesterol-depleting drug methyl-β-cyclodextrin reduced SHFV titers more than 10-fold, and subsequent addition of exogenous soluble cholesterol restored the titers to untreated-control levels (Fig. 6E). This observation has also been made for PRRSV and EAV (27, 45) and suggests that SHFV cell entry is dependent on cholesterol, which is known to inhibit both CME and caveolar/raft-mediated pathways (46).
In summary, our data indicate that endocytosis, possibly reflecting a process reminiscent of CME, is the predominant pathway by which SHFV gains entry into MA-104 cells.
Cathepsins L and B do not play a role in SHFV cell entry and replication.
Concomitant with endocytosis, some viruses require cathepsin L- and/or B-mediated cleavage of viral surface glycoproteins before, during, or after fusion of the endosome with lysosomes for infectivity (47–51). To evaluate whether SHFV entry/replication is also dependent on cathepsins, we treated MA-104 cells with increasing concentrations of the irreversible cathepsin L and B inhibitor E-64d, the cathepsin L inhibitor FYdmk, or the cathepsin B inhibitor CA-074. Whereas treatment of MA-104 cells with E-64d and FYdmk reduced infection with the nidovirus MERS-CoV, as described previously (49, 51), no effect was observed on SHFV particle production with any of the three compounds (Fig. 7A to C).
FIG 7.
Cathepsins L and B do not play a role in SHFV cell entry and replication. (A to C) Effects of pretreatment of MA-104 cells with increasing concentrations of cathepsin inhibitors on SHFV and MERS-CoV (positive control) particle yield, as determined by plaque assay. MA-104 cells were pretreated with E-64d (cathepsin L and B inhibitor), FYdmk (cathepsin L inhibitor), or CA-074 (cathepsin B inhibitor) for 4 h and then infected with SHFV or MERS-CoV at an MOI of 5. The error bars indicate the standard deviations of triplicate samples from one of two independent experiments.
SHFV particles use a proteinaceous cell surface receptor to gain entry into target cells.
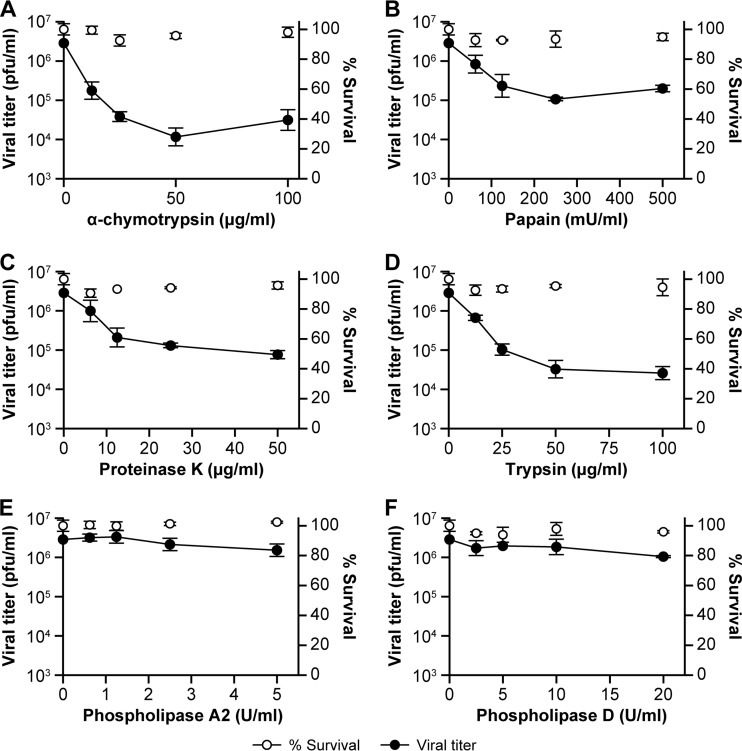
The known receptors of mammalian viruses are predominantly cell surface proteins. To evaluate whether SHFV particle cell entry is dependent on a cell surface protein, we treated MA-104 cells with increasing concentrations of the four proteases α-chymotrypsin, papain, proteinase K, and trypsin prior to SHFV particle exposure. All four proteases reduced SHFV particle production between 50- and 100-fold (Fig. 8A to D). We also treated MA-104 cells with increasing concentrations of phospholipase A2 or D to test the direct involvement of MA-104 plasma membrane lipids as SHFV receptors. These treatments did not influence SHFV particle production (Fig. 8E and F). Together, these results indicate that the SHFV receptor on MA-104 cells is a protein and that SHFV interaction with the receptor does not involve lipids.
FIG 8.
SHFV particles use a proteinaceous cell surface receptor to gain entry into target cells. (A to D) Effects of pretreatment of MA-104 cells with increasing concentrations of proteases on SHFV particle yield, as determined by plaque assay. (E and F) Effects of pretreatment of MA-104 cells with increasing concentrations of phospholipase A2 or D on SHFV particle yield. The error bars indicate the standard deviations of triplicate samples from one of two independent experiments.
CD163 is a crucial SHFV cell entry factor.
The protein CD163 is a crucial cell entry factor for the SHFV-related porcine pathogen PRRSV. We therefore hypothesized that grivet CD163 may be involved in SHFV particle entry into MA-104 cells. We incubated SHFV- and PRRSV-permissive MA-104 and MARC-145 cells (a direct subclone of MA-104 cells) with increasing concentrations of a specific anti-human CD163 antibody and, subsequent to washing, infected the cells with SHFV or PRRSV. As previously described (23), antibody treatment reduced PRRSV particle production. Likewise, SHFV particle production decreased by roughly 100-fold in both tested cell lines (Fig. 9A and B). These observations indicate that CD163 plays an important role in SHFV particle cell entry.
FIG 9.
CD163 is a crucial SHFV cell entry factor. (A and B) Effects of incubation of MA-104 or MARC-145 cells with increasing concentrations of human anti-CD163 or control antibody on SHFV and PRRSV particle yield, as determined by plaque assay. The error bars indicate the standard deviations of triplicate samples from one of two independent experiments.
DISCUSSION
In general, direct fusion of virions with cellular plasma membranes is independent of pH, whereas a low pH is required when virions usurp the cellular endocytosis pathway (52, 53). Treatments with drugs that disrupt cellular pH inhibited SHFV particle entry in a dose-dependent manner (Fig. 2A to D), suggesting a pH-dependent uptake mechanism. As observed for many other viruses (54), a low pH could be necessary for SHFV particles to induce conformational changes of their surface glycoproteins necessary to induce fusion of the virion membrane with the target cell membrane. It is worth noting, however, that pH disruption by lysosomotropic inhibitors, Na+/H+ antiporters, or V-type H+-ATPase inhibitors may cause off-target effects, as they can also disrupt virus receptor recycling, inhibit endosome maturation, or neutralize the _trans_-Golgi network (53, 55). Therefore, further experiments will have to be performed to further confirm the pH dependency of SHFV cell entry.
Endocytosis can be grossly differentiated into phagocytosis and pinocytosis (52, 53). Phagocytosis has been observed only in specialized cells engulfing very large particles (56, 57). As the vast majority of known viruses usurp pinocytic pathways (52, 53), we assumed that the small spherical virions (40 to 55 nm in diameter) produced by SHFV (8) enter host cells by pinocytosis. Pinocytosis can be divided into subpathways that are dependent on dynamin (clathrin-mediated endocytosis, caveola-mediated endocytosis, and ill-characterized pathways of clathrin-independent dynamin-mediated endocytosis) and those that are not dependent on dynamin (variably referred to as macropinocytosis, i.e., via CLIC/GEEC [58]; lipid raft-mediated endocytosis; and nonclathrin/noncaveolar endocytosis) (52, 53, 59). Our experiments with the dynamin inhibitors dynasore and Dyngo-4a, as well as with a dominant-negative dynamin mutant, suggest that SHFV utilizes a dynamin-mediated pathway (Fig. 3C to F).
We suspected CME to be the main route of SHFV entry because the arteriviruses PRRSV and EAV both use the pathway (27–29) and because CME is also a popular mode of entry for many other mammalian viruses (60–63). CME typically occurs rapidly, with virions bound to target cell surfaces entering cells within minutes. Furthermore, CME is characterized morphologically by the formation of heavily coated plasma membrane indentations (clathrin-coated pits) and the formation of characteristic clathrin-coated vesicles, both of which can be identified by electron microscopy (53, 64). We identified subcellular, membrane-proximal structures in SHFV-infected cells via electron microscopy that are consistent with classical descriptions of clathrin-coated pits and clathrin-coated vesicles (65–67) (Fig. 1A). Membrane blebbing, typical of macropinocytosis, was not observed. Formation of pits and engulfment of SHFV particles occurred within minutes of exposure (Fig. 1A). Cells pretreated with the bona fide CME inhibitors became resistant to SHFV particle entry, and clathrin heavy chain knockout also inhibited virus infection (Fig. 3A, B, and H). Together, these data indicate that CME is likely the main pathway used by SHFV to enter MA-104 cells. Our experiments were limited by the facts that only one cell line, MA-104 (and its subclones), is known to be susceptible to SHFV and that the only available SHFV readout assay available is the plaque assay, which is not sensitive enough to measure subtle effects on viral titers.
Several viruses are known to use multiple pathways to infect their target cells, or they use specific, but distinct, pathways in different cell types (reviewed in reference 68). To test the possibility of SHFV using multiple pathways in MA-104 cells, we further investigated macropinocytosis and caveolin-mediated endocytosis.
Macropinocytosis is a transient, actin-dependent cellular process used to internalize fluids and membrane into large vacuoles. Morphologically, macropinocytosis is characterized by ruffling of the cell plasma membrane induced by actin and microfilament activation (52). We did not observe membrane ruffling or blebbing in our electron microscopy studies of SHFV particle entry into MA-104 cells (Fig. 1A), and cell treatment with blebbistatin, which inhibits bleb formation, did not affect SHFV replication (Fig. 4B). Although we could demonstrate the disruption of the cellular actin network of MA-104 cells after exposure to several drugs, we could not detect an influence of that disruption on SHFV particle entry (Fig. 5). Other typical inhibitors of macropinocytosis (EIPA and wortmannin) did not influence SHFV particle entry either (Fig. 4). While EIPA, blebbistatin, and wortmannin affected VACV-eGFP infection, as described previously (69), in an optical readout assay (Fig. 4), no effect was measurable in VACV-eGFP plaque assays (data not shown). This discrepancy raises the possibility that the three inhibitors may have some effect on SHFV infection and that, because of the lower sensitivity of the plaque assay and the absence of an optical readout assay for SHFV infection, we were unable to detect it. However, together with the observation that SHFV particle entry is dependent on dynamin (Fig. 3), our data indicate that the macropinocytotic pathway is highly unlikely to be a major gateway for SHFV into MA-104 cells. Our results indicating that SHFV cell entry is independent of actin (Fig. 5) further supports a CME-like route as the major SHFV cell entry pathway (70–72).
Caveola-mediated endocytosis has been identified as the major route of entry for RVFV and other viruses (44, 73). A hallmark of this pathway is its dependence on caveolin. Treatment of cells with caveola-mediated-endocytosis inhibitors did not influence SHFV particle production (Fig. 6), but the inhibitors decreased RVFV entry in an IFA, as previously described (44), suggesting that this pathway is not used as a major entry pathway by SHFV. However, it is possible that the inhibitors used have some effects on SHFV infection that could not be detected in our plaque assay. Thus, we cannot entirely rule out the caveola pathway as an entry route for SHFV. However, we could not observe caveola-like invaginations in SHFV-infected MA-104 cells (Fig. 1A), suggesting that the pathway is unlikely to play a major role in SHFV entry.
Several exotic endocytic pathways, such as the interleukin 2 (IL-2), GEEC, flotilin, and ARF6 pathways (reviewed in reference 53), were not examined in this study. Although it is unlikely given the data presented here, SHFV could possibly enter cells by means different from CME or by a unique mechanism that incorporates components of several pathways.
Overall, our data suggest that SHFV enters cells in a similar manner to EAV and PRRSV (27–29). However, while filipin III completely inhibited EAV at a concentration of 10 μM (27), we did not see any effect of the compound on SHFV particle production.
The distribution of arterivirus host cell receptors, and differences among its orthologs, could explain the predilection of arteriviruses for particular cells. Unfortunately, bona fide cell surface receptors have not yet definitely been identified for any arterivirus. However, at least two cell surface factors, the macrophage-restricted sialoadhesin CD169/Sn/Siglec-1 and the more generally distributed CD163, have been implicated in playing major roles in PRRSV cell entry (23, 24). By using various proteases to strip SHFV target cells of surface proteins, we demonstrated that SHFV is dependent on a cell surface protein for entry (Fig. 8). We hypothesized that this factor may be CD163. Indeed antibodies against CD163 inhibited both PRRSV and SHFV cell entry (Fig. 9), suggesting that CD163 plays similar roles in the entry of both viruses. Future studies will have to address whether CD163 is a true receptor for both viruses, whether other cofactors/coreceptors are involved, and which arterivirion envelope proteins engage the receptor and mediate fusion. In addition, it will be important to confirm the results described here in other cell types, possibly including primary macrophages. To achieve these goals, and to circumvent the limitations of the plaque assay for the quantification of SHFV yields, we are currently working toward the establishment of recombinant reporter viruses and a panel of SHFV-specific antibodies to perform high-content immunofluorescence assays.
ACKNOWLEDGMENTS
The content of this publication does not necessarily reflect the views or policies of the U.S. Department of the Army, the U.S. Department of Defense, or the U.S. Department of Health and Human Services or of the institutions and companies with which we are affiliated.
Y.C., E.N.P., J.G.B., V.W.-J., and J.H.K. performed this work as employees of Tunnell Government Services, Inc., N.M.D. as an employee of MRI Global, and M.G.L. as an employee of Lovelace Respiratory Research Institute, all subcontractors of Battelle Memorial Institute, and S.Y. as an employee of Battelle Memorial Institute, all under Battelle's prime contract with NIAID (contract no. HHSN272200700016I).
We thank Kay Faaberg (U.S. Department of Agriculture, National Animal Disease Center, Ames, IA) for the generous gift of MARC-145 cells. We are grateful to Laura Bollinger, Lauren Keith, and Jiro Wada of IRF-Frederick for critically editing the manuscript and creating figures. Finally, we thank the Clinical Core group of IRF-Frederick for growing, performing quality control checks on, and quantifying the various virion preparations used for this study.
REFERENCES
- 1.Faaberg KS, Balasuriya UB, Brinton MA, Gorbalenya AE, Leung FC-C, Nauwynck H, Snijder EJ, Stadejek T, Yang H, Yoo D. 2011. Family Arteriviridae, p 796–805. _In_King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom. [Google Scholar]
- 2.Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, O'Connor DH, Friedrich TC, Goldberg TL. 2011. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One 6:e19056. doi: 10.1371/journal.pone.0019056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauck M, Sibley SD, Hyeroba D, Tumukunde A, Weny G, Chapman CA, Ting N, Switzer WM, Kuhn JH, Friedrich TC, O'Connor DH, Goldberg TL. 2013. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J Virol 87:688–691. doi: 10.1128/JVI.02433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.London WT. 1977. Epizootiology, transmission and approach to prevention of fatal simian haemorrhagic fever in rhesus monkeys. Nature 268:344–345. doi: 10.1038/268344a0. [DOI] [PubMed] [Google Scholar]
- 5.Bailey AL, Lauck M, Weiler A, Sibley SD, Dinis JM, Bergman Z, Nelson CW, Correll M, Gleicher M, Hyeroba D, Tumukunde A, Weny G, Chapman C, Kuhn JH, Hughes AL, Friedrich TC, Goldberg TL, O'Connor DH. 2014. High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population. PLoS One 9:e90714. doi: 10.1371/journal.pone.0090714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shevtsova ZV. 1967. Study of the etiology of hemorrhagic fever in monkeys. Vopr Virusol 12:47–51. (In Russian.) [PubMed] [Google Scholar]
- 7.Palmer AE, Allen AM, Tauraso NM, Shelokov A. 1968. Simian hemorrhagic fever. I. Clinical and epizootiologic aspects of an outbreak among quarantined monkeys. Am J Trop Med Hyg 17:404–412. [PubMed] [Google Scholar]
- 8.Wood O, Tauraso N, Liebhaber H. 1970. Electron microscopic study of tissue cultures infected with simian haemorrhagic fever virus. J Gen Virol 7:129–136. doi: 10.1099/0022-1317-7-2-129. [DOI] [PubMed] [Google Scholar]
- 9.Sagripanti JL. 1984. The genome of simian hemorrhagic fever virus. Arch Virol 82:61–72. doi: 10.1007/BF01309368. [DOI] [PubMed] [Google Scholar]
- 10.Sagripanti JL. 1985. Polyadenylic acid sequences in the genomic RNA of the togavirus of simian hemorrhagic fever. Virology 145:350–355. doi: 10.1016/0042-6822(85)90171-0. [DOI] [PubMed] [Google Scholar]
- 11.Sagripanti JL, Zandomeni RO, Weinmann R. 1986. The cap structure of simian hemorrhagic fever virion RNA. Virology 151:146–150. doi: 10.1016/0042-6822(86)90113-3. [DOI] [PubMed] [Google Scholar]
- 12.Godeny EK, Zeng L, Smith SL, Brinton MA. 1995. Molecular characterization of the 3′ terminus of the simian hemorrhagic fever virus genome. J Virol 69:2679–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snijder EJ, Kikkert M. 2013. Arteriviruses, p 859–879. _In_Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 14.Godeny EK, de Vries AA, Wang XC, Smith SL, de Groot RJ. 1998. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J Virol 72:862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vries AA, Post SM, Raamsman MJ, Horzinek MC, Rottier PJ. 1995. The two major envelope proteins of equine arteritis virus associate into disulfide-linked heterodimers. J Virol 69:4668–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faaberg KS, Even C, Palmer GA, Plagemann PG. 1995. Disulfide bonds between two envelope proteins of lactate dehydrogenase-elevating virus are essential for viral infectivity. J Virol 69:613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C, Yoo D. 2006. The small envelope protein of porcine reproductive and respiratory syndrome virus possesses ion channel protein-like properties. Virology 355:30–43. doi: 10.1016/j.virol.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wieringa R, De Vries AA, Post SM, Rottier PJ. 2003. Intra- and intermolecular disulfide bonds of the GP2b glycoprotein of equine arteritis virus: relevance for virus assembly and infectivity. J Virol 77:12996–13004. doi: 10.1128/JVI.77.24.12996-13004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wieringa R, de Vries AA, Rottier PJ. 2003. Formation of disulfide-linked complexes between the three minor envelope glycoproteins (GP2b, GP3, and GP4) of equine arteritis virus. J Virol 77:6216–6226. doi: 10.1128/JVI.77.11.6216-6226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firth AE, Zevenhoven-Dobbe JC, Wills NM, Go YY, Balasuriya UB, Atkins JF, Snijder EJ, Posthuma CC. 2011. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol 92:1097–1106. doi: 10.1099/vir.0.029264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wissink EH, Kroese MV, van Wijk HA, Rijsewijk FA, Meulenberg JJ, Rottier PJ. 2005. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J Virol 79:12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plemper RK. 2011. Cell entry of enveloped viruses. Curr Opin Virol 1:92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch SK. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J Virol 81:7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. 2008. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J Gen Virol 89:2943–2953. doi: 10.1099/vir.0.2008/005009-0. [DOI] [PubMed] [Google Scholar]
- 25.Das PB, Dinh PX, Ansari IH, de Lima M, Osorio FA, Pattnaik AK. 2010. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J Virol 84:1731–1740. doi: 10.1128/JVI.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. 2009. The porcine reproductive and respiratory syndrome virus requires trafficking through CD163-positive early endosomes, but not late endosomes, for productive infection. Arch Virol 154:1939–1943. doi: 10.1007/s00705-009-0527-1. [DOI] [PubMed] [Google Scholar]
- 27.Nitschke M, Korte T, Tielesch C, Ter-Avetisyan G, Tunnemann G, Cardoso MC, Veit M, Herrmann A. 2008. Equine arteritis virus is delivered to an acidic compartment of host cells via clathrin-dependent endocytosis. Virology 377:248–254. doi: 10.1016/j.virol.2008.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nauwynck HJ, Duan X, Favoreel HW, Van Oostveldt P, Pensaert MB. 1999. Entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages via receptor-mediated endocytosis. J Gen Virol 80:297–305. [DOI] [PubMed] [Google Scholar]
- 29.Kreutz LC, Ackermann MR. 1996. Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res 42:137–147. doi: 10.1016/0168-1702(96)01313-5. [DOI] [PubMed] [Google Scholar]
- 30.Earl PL, Cooper N, Wyatt LS, Moss B, Carroll MW. 2001. Preparation of cell cultures and vaccinia virus stocks. Curr Protoc Mol Biol Chapter 16:Unit 16. doi: 10.1002/0471142727.mb1616s43. [DOI] [PubMed] [Google Scholar]
- 31.Mudhasani R, Tran JP, Retterer C, Radoshitzky SR, Kota KP, Altamura LA, Smith JM, Packard BZ, Kuhn JH, Costantino J, Garrison AR, Schmaljohn CS, Huang IC, Farzan M, Bavari S. 2013. IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J Virol 87:8451–8464. doi: 10.1128/JVI.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, Fischer ER, Martellaro C, Okumura A, Chang J, Scott D, Benecke AG, Katze MG, Feldmann H, Munster VJ. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci U S A 110:16598–16603. doi: 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez L, Carrasco L. 1994. Involvement of the vacuolar H(+)-ATPase in animal virus entry. J Gen Virol 75:2595–2606. doi: 10.1099/0022-1317-75-10-2595. [DOI] [PubMed] [Google Scholar]
- 34.Drose S, Altendorf K. 1997. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J Exp Biol 200:1–8. [DOI] [PubMed] [Google Scholar]
- 35.Marsh M, Helenius A. 2006. Virus entry: open sesame. Cell 124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang LH, Rothberg KG, Anderson RG. 1993. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol 123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson AE, Noel RJ, Herlihy JT, Ward WF. 1989. Phenylarsine oxide inhibition of endocytosis: effects on asialofetuin internalization. Am J Physiol 257:C182–C184. [DOI] [PubMed] [Google Scholar]
- 38.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. 2009. Dissecting dynamin's role in clathrin-mediated endocytosis. Biochem Soc Trans 37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harper CB, Popoff MR, McCluskey A, Robinson PJ, Meunier FA. 2013. Targeting membrane trafficking in infection prophylaxis: dynamin inhibitors. Trends Cell Biol 23:90–101. doi: 10.1016/j.tcb.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L, Mali P, Kim-Kiselak C, Church G. 2014. CRISPR-Cas-mediated targeted genome editing in human cells. Methods Mol Biol 1114:245–267. doi: 10.1007/978-1-62703-761-7_16. [DOI] [PubMed] [Google Scholar]
- 43.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 44.Harmon B, Schudel BR, Maar D, Kozina C, Ikegami T, Tseng CT, Negrete OA. 2012. Rift Valley fever virus strain MP-12 enters mammalian host cells via caveola-mediated endocytosis. J Virol 86:12954–12970. doi: 10.1128/JVI.02242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, Xiao S, Wang D, Luo R, Li B, Chen H, Fang L. 2011. Cellular membrane cholesterol is required for porcine reproductive and respiratory syndrome virus entry and release in MARC-145 cells. Sci China Life Sci 54:1011–1018. doi: 10.1007/s11427-011-4236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subtil A, Gaidarov I, Kobylarz K, Lampson MA, Keen JH, McGraw TE. 1999. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc Natl Acad Sci U S A 96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol 80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirato K, Kawase M, Matsuyama S. 2013. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol 87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A 102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian Z, Dominguez SR, Holmes KV. 2013. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One 8:e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercer J, Helenius A. 2009. Virus entry by macropinocytosis. Nat Cell Biol 11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 53.Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu Rev Biochem 79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 54.Vazquez-Calvo A, Saiz JC, McCullough KC, Sobrino F, Martin-Acebes MA. 2012. Acid-dependent viral entry. Virus Res 167:125–137. doi: 10.1016/j.virusres.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 55.Clague MJ, Urbe S, Aniento F, Gruenberg J. 1994. Vacuolar ATPase activity is required for endosomal carrier vesicle formation. J Biol Chem 269:21–24. [PubMed] [Google Scholar]
- 56.Clement C, Tiwari V, Scanlan PM, Valyi-Nagy T, Yue BY, Shukla D. 2006. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J Cell Biol 174:1009–1021. doi: 10.1083/jcb.200509155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghigo E, Kartenbeck J, Lien P, Pelkmans L, Capo C, Mege JL, Raoult D. 2008. Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog 4:e1000087. doi: 10.1371/journal.ppat.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lakshminarayan R, Wunder C, Becken U, Howes MT, Benzing C, Arumugam S, Sales S, Ariotti N, Chambon V, Lamaze C, Loew D, Shevchenko A, Gaus K, Parton RG, Johannes L. 2014. Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat Cell Biol 16:595–606. doi: 10.1038/ncb2970. [DOI] [PubMed] [Google Scholar]
- 59.Sun Y, Tien P. 2013. From endocytosis to membrane fusion: emerging roles of dynamin in virus entry. Crit Rev Microbiol 39:166–179. doi: 10.3109/1040841X.2012.694412. [DOI] [PubMed] [Google Scholar]
- 60.Clemente R, de la Torre JC. 2009. Cell entry of Borna disease virus follows a clathrin-mediated endocytosis pathway that requires Rab5 and microtubules. J Virol 83:10406–10416. doi: 10.1128/JVI.00990-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol 80:6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C, Zhuang X. 2008. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc Natl Acad Sci U S A 105:11790–11795. doi: 10.1073/pnas.0803711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matlin KS, Reggio H, Helenius A, Simons K. 1982. Pathway of vesicular stomatitis virus entry leading to infection. J Mol Biol 156:609–631. doi: 10.1016/0022-2836(82)90269-8. [DOI] [PubMed] [Google Scholar]
- 64.Schelhaas M. 2010. Come in and take your coat off—how host cells provide endocytosis for virus entry. Cell Microbiol 12:1378–1388. doi: 10.1111/j.1462-5822.2010.01510.x. [DOI] [PubMed] [Google Scholar]
- 65.Heuser J. 1980. Three-dimensional visualization of coated vesicle formation in fibroblasts. J Cell Biol 84:560–583. doi: 10.1083/jcb.84.3.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Musacchio A, Smith CJ, Roseman AM, Harrison SC, Kirchhausen T, Pearse BM. 1999. Functional organization of clathrin in coats: combining electron cryomicroscopy and X-ray crystallography. Mol Cell 3:761–770. doi: 10.1016/S1097-2765(01)80008-3. [DOI] [PubMed] [Google Scholar]
- 67.Brodsky FM. 2012. Diversity of clathrin function: new tricks for an old protein. Annu Rev Cell Dev Biol 28:309–336. doi: 10.1146/annurev-cellbio-101011-155716. [DOI] [PubMed] [Google Scholar]
- 68.Conner SD, Schmid SL. 2003. Regulated portals of entry into the cell. Nature 422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 69.Mercer J, Helenius A. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 70.Mooren OL, Galletta BJ, Cooper JA. 2012. Roles for actin assembly in endocytosis. Annu Rev Biochem 81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 71.Boucrot E, Saffarian S, Massol R, Kirchhausen T, Ehrlich M. 2006. Role of lipids and actin in the formation of clathrin-coated pits. Exp Cell Res 312:4036–4048. doi: 10.1016/j.yexcr.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaksonen M, Toret CP, Drubin DG. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 73.Nabi IR, Le PU. 2003. Caveolae/raft-dependent endocytosis. J Cell Biol 161:673–677. doi: 10.1083/jcb.200302028. [DOI] [PMC free article] [PubMed] [Google Scholar]