Keratin-dependent regulation of Aire and gene expression in skin tumor keratinocytes (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 31.
Published in final edited form as: Nat Genet. 2015 Jul 13;47(8):933–938. doi: 10.1038/ng.3355
Abstract
Expression of the intermediate filament protein keratin 17 (K17) is robustly upregulated in inflammatory skin diseases and in many tumors originating in stratified and pseudostratified epithelia1-3. We report that Autoimmune regulator (Aire), a transcriptional regulator, is inducibly expressed in human and mouse tumor keratinocytes in a K17-dependent manner and required for a timely onset of _Gli2_-induced skin tumorigenesis in mice. Induction of Aire mRNA in keratinocytes depends upon a functional interaction between K17 and the heterogeneous nuclear ribonucleoprotein hnRNP K4. Further, K17 colocalizes with Aire protein in the nucleus of tumor-prone keratinocytes, and each are bound to a specific promoter region featuring a NF-κB consensus sequence in a relevant subset of K17- and Aire-dependent pro-inflammatory genes. These findings provide radically new insight into keratin intermediate filament and Aire function, along with a molecular basis for the K17-dependent amplification of inflammatory and immune responses in diseased epithelia.
Keywords: Keratin 17 (Krt17), Autoimmune regulator (Aire), hnRNP K, Skin, Inflammation, Cancer
Main text
High levels of Krt17 expression have been correlated to aggressive behavior and poor prognosis for several types of human tumors2. Genetic loss of Krt17, but not the related Krt14, delays tumor onset in a Gli2tg mouse model of basaloid skin tumorigenesis, correlating with striking changes in the amplitude and character of the inflammatory and immune responses5. Moreover, Krt17 positively regulates several effectors of mitogenic signaling associated with oncogenic transformation, e.g., Akt/PKB6, mTOR7, and Rac1 GTPase8, and is necessary to sustain normal Akt/PKB activation in the context of _Gli1_-mediated oncogenic transformation in Ewing Sarcoma6. Whether Krt17 impacts additional types of tumors in a similar manner, and which mechanism(s) account for its influence, are unknown.
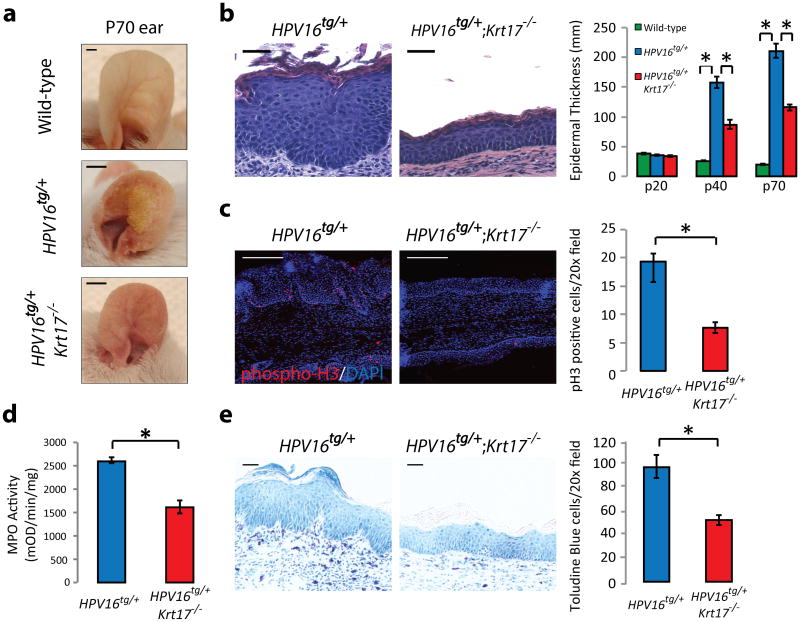
Targeted expression of the early genes from HPV type 16 (E1 to E7) to basal keratinocytes of epidermis causes tumors in adult mouse skin that resemble human skin squamous cell carcinoma9. Tumors arise with complete penetrance between postnatal day 60 (P60) and P120 in HPV16tg/+ ear skin (FVB/N strain) (Supplementary Fig. 1a), with no apparent gender discordance. While normally restricted to ectodermal appendages and glabrous skin10, expression of K17 is robustly upregulated in the interfollicular epidermis between P20 and P40 in HPV16tg/+ mice (Supplementary Fig. 1b). Relative to HPV16tg/+;Krt17+/+, HPV16tg/+;Krt17-/- mice exhibit delayed hyperplasia and tumorigenesis in ear skin (Fig. 1a-b, Supplementary Fig. 1c). As had been observed in Gli2tg/+;Krt17-/- mice5, the delay correlates with profound reductions in key determinants of tumor growth including mitotic activity (Fig. 1c), blood vessel expansion (Supplementary Fig. 1d), inflammatory and immune response readouts (Supplementary Fig. 1e) including myeloperoxidase activity reflecting neutrophil infiltration (Fig. 1d), and dermal mast cell density (Fig. 1e). Transgene expression, p53 levels, and apoptosis are indistinguishable between genotypes in ear skin (Supplementary Fig. 1f-h). There is no indication of cell fragility in HPV16tg/+; Krt17-/- skin by electron microscopy (Supplementary Fig. 1i). Commercial qRT-PCR arrays identified many pro-inflammatory cytokines as being significantly depressed in tumor-prone ear tissue from HPV16tg/+;Krt17-/- relative to controls (Supplementary Tables 1-5). Thus, the loss of Krt17 delays tumorigenesis in mouse skin and significantly attenuates the expression of several key pro-inflammatory signaling molecules in two oncogenic paradigms (Gli2tg and HPV16tg) involving strains that are differentially sensitive to skin carcinogenesis (C57Bl/6 and FVB/N)11.
Figure 1. Loss of Krt17 attenuates _HPV16tg_-induced skin tumorigenesis.
a) Macroscopic images of P70 ears from wild-type, HPV16tg/+, or HPV16tg/+;Krt17-/- mice. Scale bars = 1 cm. b) H&E staining of P70 mouse ear tissue sections and quantitation of average epidermal thickness at P20 (n = 51), P40 (n = 54), and P70 (n = 28) across genotypes. n = number of biological replicates. Scale bars = 20 μm. Error bars are s.e.m. c) Immunostaining and quantitation (n = 6 biological replicates) of phospho-Histone H3 (red) in P70 mouse ear tissue sections. Hoescht DNA stain (blue). Scale bars = 20 μm. Error bars are s.e.m. d) Myeloperoxidase (MPO) activity assay for neutrophil activation (n = 3 biological replicates, each with 4 techinical replicates). Error bars are s.e.m. e) Toluidine blue staining and quantitation (n = 6 biological replicates) of mast cells in P70 ear tissue sections. *p<0.05. Scale bars = 20 μm. Error bars are s.e.m.
To explore how pro-inflammatory gene expression may be regulated by K17, we devised a custom, 96-well plate based, real-time PCR (qRT-PCR) assay to quantitate mRNA levels for inflammation- and disease-relevant genes (Supplementary Table 6) in A431 cells (derived from a human epidermoid carcinoma and K17-expressing1). Twenty-two genes were consistently upregulated (>2-fold) in A431 cells following treatment with TPA (Supplemental Fig. 2a), which elicits a robust inflammatory response in keratinocytes12. Of those, nineteen are similarly upregulated in P40 HPV16tg/+ ear tissue (Fig. 2a). Compared to non-silencing shRNA control, A431 cells with stable KRT17 knockdown13 exhibit a significantly attenuated response to TPA (Supplemental Fig. 2a). Re-introduction of K17, but not the highly homologous K4214, largely restored TPA-dependent cytokine upregulation in A431 cells (Supplementary Fig. 2b). Therefore, the expression of multiple pro-inflammatory cytokines also depends on KRT17 in human skin tumor keratinocytes.
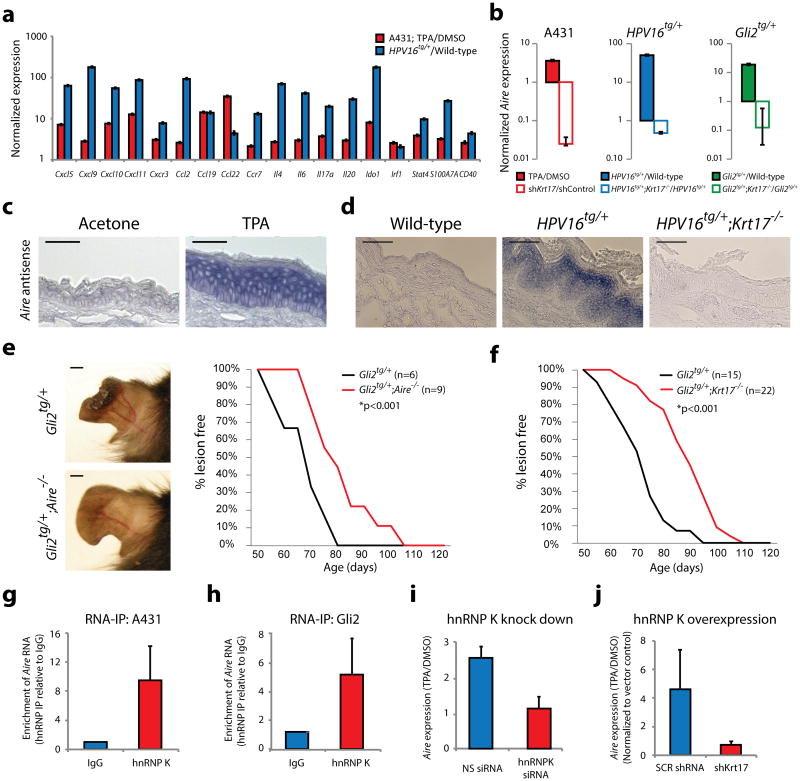
Figure 2. Aire expression, function, and regulation in skin tumor keratinocytes.
a) Normalized expression for 18 of 19 gene transcripts common to A431 (red, n = 5 biological replicates) and HPV16tg/+ (blue, n = 6 biological replicates) paradigms. Error bars are s.e.m. b) Normalized expression for Aire (the 19th gene target) transcript levels in A431 (red), HPV16tg/+ (blue) and Gli2tg/+ (green) paradigms with (solid bars) or without (open bars) Krt17 expression. Error bars are s.e.m. c) Aire RNA in situ hybridization (ISH) in wild-type FVB/N mouse ear tissue sections treated with acetone or TPA. Scale bars = 10 μm. d) Aire ISH in ear tissue sections from age-matched wild-type, HPV16tg/+, and HPV16tg/+;Krt17-/- mice. Scale bars = 20 μm. e) P80 mouse ears from Gli2tg/+ and Gli2tg/+;Aire-/- littermates. Scale bars = 1 cm. Graph depicts percent Gli2tg/+;Aire-/- (n = 9) mice lesion-free over time relative to Gli2tg/+ littermates (n = 6). f) Graph depicts percent of Gli2tg/+;Krt17-/- mice (n = 22) lesion-free over time relative to Gli2tg/+ littermates (n = 15). g-h) Enrichment of Aire transcript with hnRNP K immunoprecipitation (RNA-IP) relative to IgG control in A431 keratinocytes (n = 9 biological replicates) and Gli2tg/+ keratinocytes in primary culture (n = 9 biological replicates). Error bars are s.e.m. i) TPA-induced Aire transcript levels in A431 keratinocytes expressing non-silencing (NS) or hnRNPK-targeting siRNA oligos (n = 7 biological replicates). Error bars are s.e.m. j) TPA-induced Aire transcript levels in A431 keratinocytes overexpressing hnRNP K while stably expressing shRNA targeting Krt17 (sh_Krt17_), relative to scrambled (SCR) sequence (n = 3 biological replicates). Error bars are s.e.m.
Several observations converged on Aire (Autoimmune regulator), a transcriptional regulator with a well-known role in the medullary thymus towards the establishment of tolerance15-17, as a component of the K17-mediated regulation of gene expression in skin keratinocytes. First, mutation in AIRE causes autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED)18, which often presents with ectodermal anomalies in tissues that express Krt1710 (e.g., alopecia, nail dystrophy, vitiligo, enamel hypoplasia). Second, several of the pro-inflammatory genes determined to be _Krt17_-dependent in tumor-prone keratinocytes (Supplemental Fig. 2a) are _Aire_-dependent in thymic epithelial cells (e.g., Ccl19, Ccl22, Cxcl5, Ccr7, Il6, Cd40)19-21. Third, qRT-PCR and/or RNA in situ hybridization show that Aire mRNA expression is induced and _Krt17_-dependent in several settings including TPA-treated human A431 cells (Fig. 2b), ear tissue from P40 HPV16tg/+ (Fig. 2b,d) and P70 Gli2tg/+ mice (Fig. 2b), and TPA-treated ear skin of C57bl/6 and FVB/N wild-type mice (Fig. 2c, Supplemental Fig. 2c). Sense strand controls yielded negative findings (Supplemental Fig. 2d-e). Aire expression is low in untreated or wild-type skin (Fig. 2c-d)22. The observed levels of induced Aire mRNA in treated or diseased skin remain below constitutive levels in the thymus (Supplemental Fig. 2c). Fourth, there is a report of a physical interaction between Aire and K17 proteins23, which we confirmed (Supplementary Fig. 2f). Finally, we crossed Aire-/- with Gli2tg/+ mice (both available in the C57Bl/6 strain) to assess the impact of Aire loss on skin tumorigenesis (Fig. 2e). The average age of tumor onset was 79 ± 3.7 (s.e.m.) days in male Gli2tg/+;Aire-/- mice, compared to 65 ± 3.9 days in Gli2tg/+ mice (p<0.001). A similar delay was previously reported for Gli2tg/+;Krt17-/- mice5 (Fig. 2f).
Importantly, Aire expression is not _Krt17_-dependent in the medullary thymus (Supplemental Fig. 3a-b), which expresses Krt1710. Further, loss of Krt17 does not induce a break in central tolerance leading to systemic autoimmunity, unlike the case for Aire loss24 (Supplemental Fig. 3c-e). Our findings thus reveal a novel and significant role for extra-thymic Aire expression in keratinocytes, particularly during skin tumorigenesis, where its expression depends upon Krt17.
We next investigated how Aire mRNA levels depend upon K17. The ribonucleoprotein hnRNP K, a versatile regulator of gene expression with a known role during tumorigenesis25, physically and functionally interacts with K17 to impact the expression of the Cxcr3 ligands Cxcl9, Cxcl10, and Cxcl11 along with several other cytokines in tumor skin keratinocytes4. We report here that the Aire mRNA is also bound to hnRNP K, in a TPA- as well as K17-dependent manner, in human A431 and in mouse Gli2tg keratinocytes (Fig. 2g-h). The TPA-dependent induction of Aire mRNA is markedly attenuated following siRNA-mediated hnRNP K knockdown in A431 keratinocytes (Fig. 2i). Overexpression of hnRNP K suffices to induce Aire levels in A431 cells, again in a _Krt17_-dependent manner (Fig. 2j). Thus, Aire mRNA transcripts are regulated in a K17-and hnRNP K-dependent manner in skin tumor keratinocytes.
We next assessed whether K17 also regulates Aire at the protein level. Consistent with previous reports26, 27, transfected Aire fusion proteins frequently localize to round and smooth surfaced punctae in the nucleus and cytoplasm of human A431 cells (Fig. 3a) and mouse keratinocytes (data not shown) in the absence of stimulus. These Aire-positive punctae are distinct from lysosomes or stress granules, but occur immediately adjacent nuclear PML bodies (Supplemental Fig. 4a), which are associated with the storage of proteins poised for transcriptional activation28. Further, heat shock protein 70 (HSP70), previously identified to associate with Aire29 and keratin30, colocalizes with the Aire-positive nuclear punctae and does so in a _Krt17_-dependent manner (Supplemental Fig. 4b). Strikingly, within 1 hr of TPA treatment, the nuclear localization pattern of Aire shifts from punctate to diffuse (Fig. 3a). This shift is markedly hindered in sh_Krt17_-expressing A431 lines (Fig. 3a), though the punctae eventually diffuse later (6-12 hr; data not shown). Collectively, these findings indicate that K17 physically interacts with Aire and impacts its sub-nuclear distribution in response to TPA (a process that may require HSP70 association), towards the induction of target gene expression.
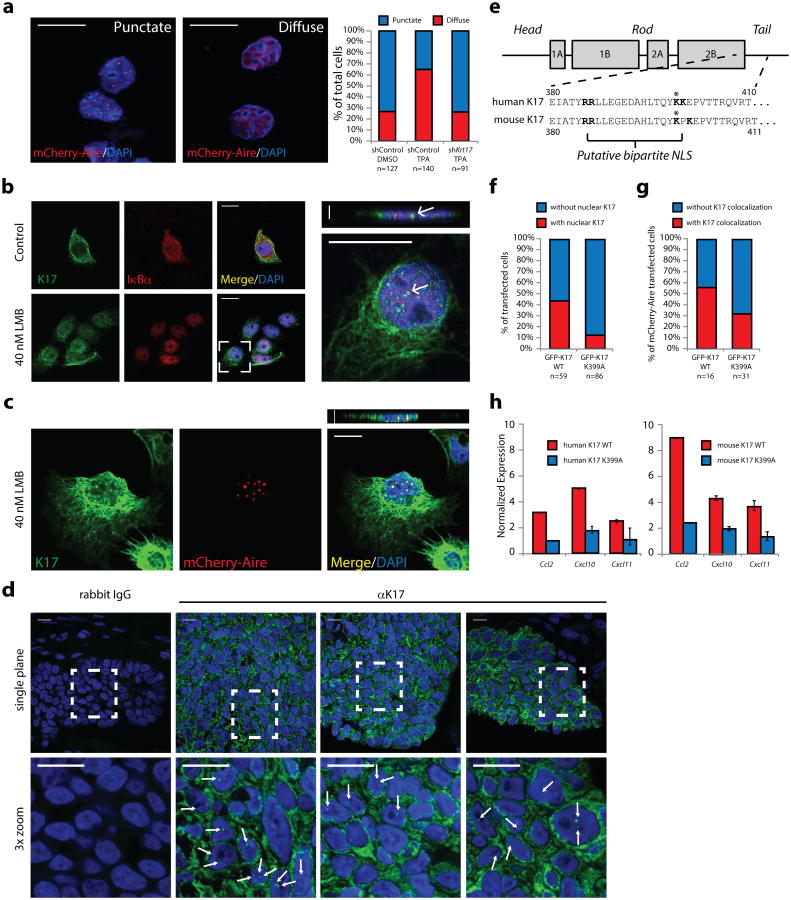
Figure 3. K17 regulates Aire subnuclear distribution and localizes to the nucleus to promote gene expression.
a) Apotome-acquired images of subnuclear distribution of mCherry-Aire in A431 keratinocytes. Graph indicates percentage of cells showing punctate (blue) or diffuse (red) pattern following TPA treatment, relative to DMSO. sh_Krt17_, cells stably expressing shRNA targeting Krt17. n = number of cells analyzed. b) Single-plane confocal images of control- or leptomycin (LMB)-treated A431 keratinocytes immunostained for K17 and IκBα (positive control for LMB treatment). Inset at right highlights K17-positive nuclear punctae (arrows) and includes z-plane image. c) Same as in 3b, except A431 cells were transfected with mCherry-Aire prior to treatment and K17 immunostaining. Images in a, b, and c are representative from 15, 10, and 5 distinct experiments, respectively. Scale bars = 5 μM (a-c), 1 μm (z-planes). d) Single-plane confocal images of K17 immunostaining (or rabbit IgG control) in tissue sections of human skin basal cell carcinoma. Bottom frames, 3× digital zooms of boxed regions in top row. Arrows denote K17-positive nuclear punctae. Scale bars = 20 μm. e) Schematic of keratin protein highlighting a conserved, predicted, bipartite nuclear localization sequence (NLS) (in bold letters). Asterisk denotes Lys399. f) Graph depicting percent of HeLa cells with nuclear punctae positive for GFP-K17, wild-type (WT) or K399A (NLS mutant), as observed by confocal microscopy. n = number of cells. g) Graph depicting percent of A431 cells where mCherry-Aire punctae colocalize with GFP-K17 WT or GFP-K17 K399A. n = number of mCherry-Aire positive cells counted. h) Normalized expression of TPA-induced target gene transcript levels in A431 Krt17 null keratinocytes transfected with GFP-K17 WT or GFP-K17 K399A, either human (left) or mouse (right) species. n = three biological replicates. Error bars are s.e.m.
We next asked whether K17 might also localize to the nucleus (along with Aire) in keratinocytes. In the presence of leptomycin B (LMB), which inhibits exportin 1-dependent nuclear export (thereby trapping low abundancy or rapidly-shuttling proteins31), K17 occurs in the form of intra-nuclear punctae in human A431 keratinocytes (Fig. 3b), where it readily colocalizes with transfected Aire protein (Fig. 3c), and in HeLa cells (Supplemental Fig. 4c-d). Intra-nuclear K17 also occurs in LMB-treated mouse epidermal keratinocytes in primary culture, where it colocalizes with type II partner K5 (Supplemental Fig. 4e). No evidence of nuclear (or any form of) K17 could be detected in LMB-treated Krt17-/- mouse keratinocytes (Supplemental Fig. 4f), establishing the specificity of our reagents. Occurrence of K17 and Aire in the nucleus is further supported by western blot analyses performed on subcellular fractions from non-LMB treated cells, including one enriched for nucleoplasmic proteins (e.g., Histone H3) and devoid of nuclear envelope markers (Supplemental Fig. 4g). By contrast K14 or K18, which also occur in A431 keratinocytes, do not yield the same fractionation pattern and have not been observed in the nucleus under these assay conditions (Supplementary Fig. 4h). Furthermore, nuclear K17 can be readily observed in tumor keratinocytes in tissue sections prepared from biopsies of skin basal cell carcinomas from patients (Fig. 3d). Bioinformatic analyses revealed a putative nuclear localization sequence (NLS) within the K17 sequence32 (Fig. 3e), a dilysine motif in the tail domain, which is conserved across species for K17, but not present in other type I intermediate filament proteins. Relative to wild-type K17, transient transfection of GFP-K17 (either human or mouse sequence) harboring a single point mutation within the putative NLS (p.Lys399Ala) into Krt17-/- A431 keratinocytes (see methods) is sufficient to attenuate the ability of K17 to: i) localize to the nucleus (Fig. 3f); ii) co-localize with nuclear mCherry-Aire punctae (Fig. 3g), and iii) foster a full cytokine expression response following TPA (Fig. 3h). These findings establish that K17 can occur in the nucleus, and suggest that the nuclear form of K17 may play a role in the rapid dispersion of the normally quiescent Aire-containing nuclear punctae following a relevant stimulus.
Though not believed to directly bind DNA33, Aire binds a plethora of transcriptional proteins to activate target gene expression20, 31. Chromatin immunoprecipitation (ChIP) assays were conducted to assess whether Aire protein, along with K17, each occur at the promoter of relevant inflammatory and immune response genes in tumor-prone keratinocytes (see Supplementary Table 7). Using RFP-trap beads in mCherry-Aire transfected A431 cells (with mCherry vector as control), ChIP analysis revealed enrichment for specific segments within the proximal 5′ upstream region for MMP9, CCL19, CXCL10 and CXCL11, but not control, genes (Fig. 4a-b). Endogenous K17 can be readily immunoprecipitated from nuclear fractions prepared from A431 cells, whether TPA-treated or not (Supplemental Fig. 5a). Following TPA, specifically, the same or adjacent segments within the MMP9, CCL19, CXCL10, and CXCL11 gene promoters are enriched in K17 IPs (Fig. 4c-d). No enrichment was observed after DMSO treatment or with pre-immune serum IP. We verified that PCR-amplified promoter fragments migrate at the expected size upon gel electrophoresis (Supplementary Fig. 5b). These ChIP findings indicate K17 and Aire each associate with the same promoter regions of select _Krt17_-dependent target genes.
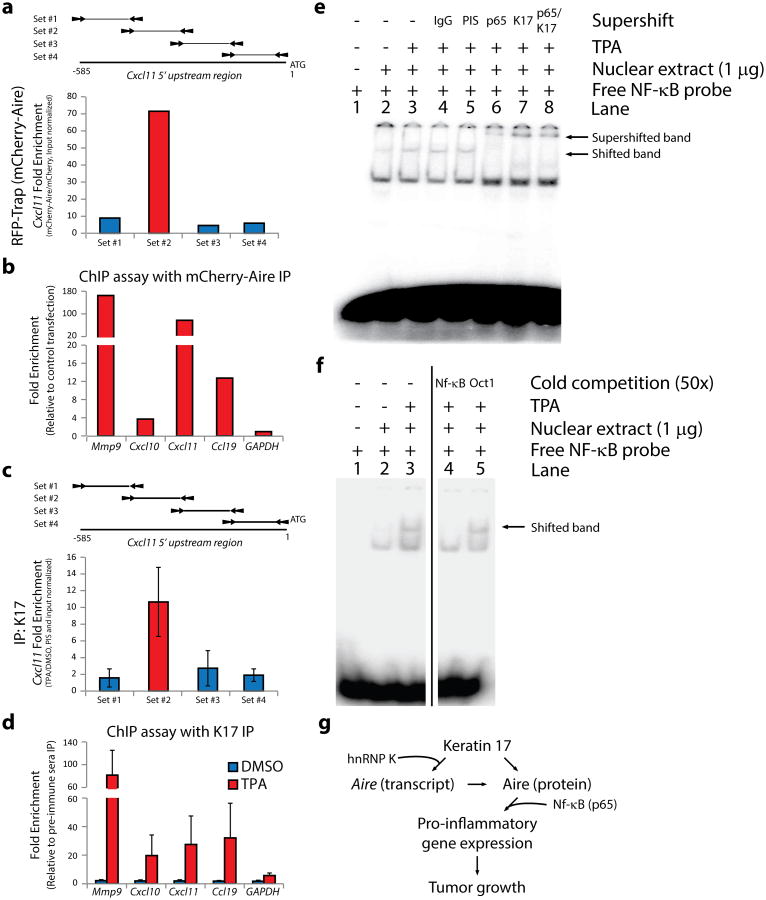
Figure 4. K17 and Aire associate with target gene promoter regions.
a) Representative ChIP assay from A431 keratinocytes depicting enrichment of a specific 5′upstream sequence of CXCL11 transcript following mCherry-Aire immunoprecipitation, relative to unfused mCherry. Schematic of the 5′upstream region for Cxcl11 with locations of qRT-PCR primer sets is provided. b) Summary of ChIP assays depicting enrichment of single DNA segments within the 5′upstream sequence of select genes with mCherry-Aire IP relative to unfused mCherry. c) Summary of ChIP assays using K17 IP, relative to pre-immune sera (PIS) control, depicting TPA-induced enrichment of the same 5′upstream sequence of the CXCL11 transcript as in Fig. 4a. n = seven biological replicates. Error bars are s.e.m. d) Summary of ChIP assays using K17 IP, relative to pre-immune sera control, depicting the TPA-induced enrichment of single DNA segments within the 5′upstream sequence of select genes. n=7 biological replicates. Error bars are s.e.m. e) EMSA analysis of radiolabeled NF-κB consensus oligonucleotides using nuclear extracts from A431 keratinocytes treated with TPA, relative to DMSO control. Supershift analysis was conducted with antibodies against p65 (lane 6) and K17 (lane 7), relative to anti-IgG (lane 4) and PIS (lane 5) controls. Image represents 1 of 5 biological replicates. f) Cold competition EMSA analysis using nuclear extracts and radiolabeled oligonucleotide as in e) with inclusion of 50-fold excess non-labeled oligonucleotide (lanes 4-5). Image represents 1 of 3 biological replicates. g) K17 regulates Aire at both the transcript and protein levels to promote inflammatory gene expression and skin tumorigenesis.
Analysis of the promoter segments enriched in ChIP samples for mCherry-Aire and K17 revealed the presence, in all cases, of consensus binding sites for NF-κB (5′-GGGRNNYYCC-3′)34 (Supplemental Fig. 5c-d). NF-κB plays a keratinocyte-autonomous role in skin inflammatory conditions35, and thus represents an ideal candidate to mediate the recruitment of K17 to relevant gene promoters. Electrophoretic mobility shift assays confirm that a protein complex associates with a NF-κB consensus oligonucleotide probe in a TPA-dependent manner in nuclear extracts prepared from A431 keratinocytes (Fig. 4e-f). Adding antisera against either K17 or the NF-κB subunit p65 yields a supershift in the mobility of the NF-κB probe, only in the presence of TPA (Fig. 4e). The use of pre-immune sera or anti-IgG control antibodies (Fig. 4e), excess unlabeled oligonucleotide as competitor (Fig. 4f), or non-TPA induced Oct1 oligonucleotide (Supplemental Fig. 5e) confirms specificity. Lastly, reciprocal co-IPs indicate that endogenous K17 and p65 interact in A431 keratinocytes (Supplemental Fig. 5f). p65/NF-κB thus may act as a molecular bridge between K17 and Aire at promoter sequences for the specific set of genes they co-regulate in tumor-prone skin keratinocytes.
In concluding, the findings reported here establish a novel role for K17 in regulating gene expression at the transcriptional level in skin keratinocytes, which involves a nuclear-localized form of this keratin protein (Fig. 4g). They provide a mechanistic basis for a positive feedback loop whereby K17 upregulation, an early event when inflammation sets in, would promote the maintenance of a specific type of pro-inflammatory and immune response in skin. Lastly, they also establish the involvement of Aire in promoting gene expression in keratinocytes and skin undergoing acute inflammation or tumorigenesis, a newly defined role that requires a physical and functional partnership with K17.
Online Methods
Mouse Models
All protocols involving mice were approved by the Johns Hopkins Institutional Care & Use Committee. C57BL/6 Krt17-/- mice36 were bred to the HPV16tg/+ transgenic mice (FVB/N strain, obtained from the National Cancer Institute) to create HPV16 tg/+;Krt17-/- mice. The resulting HPV16 tg/+;Krt17+/- mice were back-crossed with wildtype FVB/N mice for at least 6 generations. To generate Gli2tg/+;Aire-/- mice, C57Bl/6 Aire+/- mice were obtained from Jackson Laboratories (stock #006360) and bred with C57Bl/6 Gli2tg/+ mice; Gli2tg/+;Aire+/- mice were crossed with Aire+/- littermates to generate the progeny of interest. All mice with tumor transgenes were housed individually or with littermate controls upon weaning, and fed rodent chow and water ad libitum. Both male and female mice were used in this study. The sample size of mice required for this study was empirically determined from previous experience5. Mice deemed sickly by veterinary staff were not utilized. Investigator blinding and randomization were not conducted. Genotyping protocols have been previously described for the _Gli2tg_5, Krt175, HPV16tg (National Cancer Institute Mouse Repository, strain code #01XT3), and Aire alleles (Jackson Laboratories, stock #006360).
Cell lines
Mouse epidermal keratinocytes were isolated for primary culture or immortalized as described37. Parental A431 and HeLa cell lines (ATCC) were confirmed to be mycoplasma free (data not shown). Generation of A431 cells stably expressing sh_Krt17_ has been reported13. Generation of A431 cells null for K17 was conducted by CRISPR-Cas9-mediated genome engineering38. Specifically, a target sequence in the first exon of human KRT17 (5′-GGCTCCTCCGGCCTGGGGGGCGG-3′ (PAM motif underlined)) was chosen and a 20-nucleotide guide sequence (5′-GGCTCCTCCGGCCTGGGGGG-3′) was cloned into the BbsI site of pX458 (pSpCas9(BB)-2A-GFP; obtained from Addgene (plasmid #48138)) according to the cloning protocol38. For sequencing purpose, targeted K17 regions in the knockout cell line were PCR amplified and cloned into a pJET1.2/blunt cloning vector using the CloneJET™ PCR Cloning Kit (Life Technologies, #K1231). Sequencing results showed a frameshift and premature stop codon formation at both alleles of K17 (data not shown).
Antibodies, reagents, and plasmid constructs
The primary antibodies utilized in this study included: rabbit polyclonal against Krt1710, phospho-histone H3 (Cell Signaling #9701), Histone H3 (Cell Signaling #9715), mCherry (BioVision #5993), p65 (Santa Cruz #SC-372X), and Nesprin-3 (GeneTex #GTX87974); goat polyclonal against eIF3η (Santa Cruz #SC-13677) and p53 (Santa Cruz #SC-315); chicken polyclonal against Krt14 (Covance #AF64) and Krt5 (Covance #AF138); rat polyclonal against CD11b (eBiosciences #11-0112-81), CD207 (eBiosciences #14-2075-82), CD4 (BD Biosciences #550280), and CD45 (eBiosciences #45-0451-80); hamster polyclonal against CD11c (eBiosciences #17-0114-81); and mouse monoclonal against PECAM-1 (Chemicon #CBL1337), Hsp70 (StressGen #ADI-SPA-812), LAMP-1 (DSHB, University of Iowa #H4A3), Actin (Sigma #A5441), GAPDH (Santa Cruz #SC-365062), IκBα (Cell Signaling #4814), E7 (Invitrogen #28-0006), hnRNP K (Santa Cruz #SC-32307) and PML (Santa Cruz #SC-966). Secondary antibodies utilized include Alexa 488, Alexa 594, and Alexa 647 (Invitrogen) for indirect immunofluorescence, and horseradish peroxidase conjugated goat-anti-mouse, goat-anti-rabbit, and rabbit-anti- goat (Sigma) for chemiluminescence Western blotting. DNA was immunostained using Hoescht (Sigma). All commercial antibodies were used according to manufacturer's recommendation. TUNEL (Roche #11767291910) staining for apoptotic cells was done according to the manufacturer's instruction. TPA (Sigma #P1585) was dissolved in DMSO (for cell treatments) or acetone (for ear tissue topical treatment) and used at 200 nM or 25 ng/mL working concentration, respectively. Leptomycin B (LMB; Sigma #L2913) was dissolved in 70% methanol and used at 40.7 nM working concentration.
A full-length human Aire cDNA clone (a gift from P. Peterson) was moved into a mCherry-C1 vector (Clontech) using HindIII and SacII restriction enzymes, generating mCherry-Aire. mCherry and mCherry-Aire plasmids were transiently transfected into A431 cells using FuGene HD (Promega #E2311)) at a 1:3 (DNA amount:FuGene volume) ratio according to manufacturer's instruction. siRNA oligonucleotides targeting hnRNP K and their transfection protocol have been previously described4. Overexpression of hnRNP K from plasmid has been described4. The GFP-K17 K399A mutant was generated from pEGFP-C3-K17 wild-type plasmid using the Phusion mutagenesis kit (Life Technologies #F-541).
Tissue harvesting and morphological analyses
Histological assays for all ear tissue sections5 and electron microscopy39 were conducted as described. Epidermal thickness measurements, image quantitation, and myeloperoxidase assay were done as described5, 39. Basal cell aspect ratio was determined by dividing length and width of individual basal cells using the ImageJ software. Immunohistochemistry and immunofluorescence images were acquired using a Zeiss fluorescence microscope with Apotome attachment. Confocal immunofluorescence images were acquired using a Zeiss LSM 710. Images from like experiments were equally brightened, contrasted, and cropped using ImageJ software for optimal presentation.
For analysis of autoinflammation, all tissues were obtained from male mice between 4-10 months of age (n=4-5, per genotype). Tissues were embedded longitudinally in paraffin, and 5-μm sections were cut and stained with H&E or Masson's Trichrome Blue (Histoserv) prior to microscopic evaluation and grading. Grading was performed by two independent blinded investigators and averaged.
De-identified samples from paraffin embedded blocks of diagnostic biopsies for human basal cell carcinomas were used for immunofluorescence analysis also under Hopkins IRB protocol approval (NA_00072381). Tissue sections were immunostained for K17 and Hoechst and images were acquired with Apotome attachment as stated above.
Flow cytometry
Mouse ears were harvested, washed with PBS, split into halves, and placed in 0.4 mg/mL Liberase (Roche #05-401-054-001) diluted in serum-free RPMI 1640 containing 5% penicillin/streptomycin for 1.5 hr at 37°C. Liberase was inactivated by adding complete RPMI 1640 containing 5% FCS and tissues were manually homogenized by syringe prior to passage through a 70 μm-pore filter and centrifugation at 200 × g for 8 min. Cells were resuspended in PBS and centrifuged in a 30%-70% Percoll gradient (GE Healthcare #17-0891-01). Mononuclear cells at the gradient interface were collected, washed, resuspended in PBS, incubated with 5 μg/mL FcR blocker (anti-CD16/CD32, eBioscience), washed with PBS, and resuspended in staining buffer (1% fetal calf serum in PBS) prior to labeling with respective antibodies for 30 min on ice. After washing twice with staining buffer, cells were collected by FACS Calibur (BD Biosciences) and analyzed using FlowJo (Tree Star).
RNA in situ hybridization
Aire sense and antisense probes were generated by PCR subcloning a 418 bp fragment from a mouse Aire cDNA plasmid40 (provided by P. Peterson, Tartu University, Estonia) into the pCR II-TOPO vector (forward primer: 5′-AAGAAGCCAGATGGCAACTT-3′; reverse primer: 5′-ACACGGCACACTCATCCTCG-3′. In vitro transcription using T7 (Ambion #2082) and Sp6 (Ambion #2071) polymerases yielded sense and antisense probes, which were labeled with DIG and purified using illustra microspin G-50 columns (GE Healthcare #27-5330-01). 5 μL of probe was used per slide. For TPA treatment, one mouse ear was treated four times, 48-hrs apart from each other, with 25 ng/mL TPA, with the other ear of the same mouse treated with acetone as a control. ISH protocol was conducted as previously reported. The time of alkaline phosphatase reaction was the same for all samples. Four sets of biological replicates were utilized, with one representative being depicted.
Protein extraction, subcellular fractionation, and co-immunoprecipitation
Protein lysates for Western blotting were prepared in urea sample buffer (8 M deionized urea, 0.5% SDS, 30 mM Tris pH6.8, 5% glycerol, 5% β-ME) after homogenization (ear tissue) or PBS washing (cultured cells). All samples were sheared using progressive gauged needles (22 ½, 25 ½, 26 ½) and subjected to a Bradford assay and processed for Western blotting analysis as previously described5. Subcellular fractionation was conducted based on correspondence with R. Foisner (Max F. Perutz Laboratories). After 3× PBS rinsing on ice, cells were scraped into a hypotonic lysis buffer (10 mM Tris pH7.5, 1 mM MgCl2, 10 mM KCl plus protease inhibitors), incubated on ice for 10 minutes, and sheared with 20½ gauge needle to release nuclei. After spinning down at 3000 rpm for 5 minutes, supernatant was removed (soluble fraction) and pelleted nuclei were resuspended in nuclear envelope extraction buffer (20 mM HEPES pH7.9, 420 mM NaCl, 1% Triton-X 100, 1.5 mM MgCl2, 0.2 mM EDTA plus protease inhibitors), and twice incubated on ice for 10 min followed by 1 min vortex prior to spinning at 13,000 rpm for 10 minutes. The supernatant containing nuclear envelope (NE) was retained. The pellet containing histones (Chr) was resuspended in urea sample buffer. Sol, NE, and Chr samples were electrophoresed on 10% SDS-PAGE and subjected to Western blot analysis using relevant markers.
Immunoprecipitation for Krt17, p65, and hnRNP K was conducted as previously described4, 34. mCherry and mCherry-Aire were immunoprecipitated using 10 μL RFP-trap beads (Chromotek) per sample incubated for 1 hr at 4°C with rotation. HPV16 E7 protein was immunoprecipitated from P2 mouse skin subjected to the Dynabeads co-immunoprecipitation kit (Invitrogen) containing protease inhibitors. Protein content was determined by Bradford assay, and 1.5 mg of HPV16 E7 antibody or rabbit IgG was coupled to Dynabeads which, after clearing, were incubated with extracted proteins for 1 hr at 4°C. After washing and elution, IP samples were then electrophoresed by SDS-PAGE and subjected to Western blot analysis. RNA-immunoprecipitation was conducted as previously described4.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Commercial qRT-PCR plates (SABiosciences; #PAMM-011A, #PAMM-025, #PAMM-073, #PAMM-014A, and #PAMM-052) were performed on ear tissue samples from P40 HPV16tg/+ and HPV16tg/+;K17-/- mice. Custom qRT-PCR assays were performed on ear tissue samples from P80 Gli2tg/+ and Gli2tg/+;Krt17-/-, P40 HPV16tg/+ and HPV16tg/+;K17-/- mice, and A431 cells stably expressing control or sh_Krt17_ plasmids. Total RNA from ear tissue was isolated using Trizol (Invitrogen), followed by DNAseI treatment (Qiagen) and cleanup on RNeasy Mini column (Qiagen). RNA from A431 cells was isolated using the RNEasy kit (Qiagen). All RNA concentration and purity was assessed by spectrophotometry. 1 μg of RNA was reverse-transcribed with the RT2 First strand Kit (Qiagen) or iScript (BioRad). qRT-PCR was performed on the first strand cDNA using the RT2 Profiler PCR Array (Qiagen) or SSO-Advanced SYBR green (BioRad) as described by the manufacturer. PCR parameters for custom qRT-PCR screen = 95°C for 5 minutes, followed by 40 cycles of 95°C for 10 sec + 55°C for 30 sec. No template controls, no reverse-transcriptase controls, standard curves, and a melt curve were included with every PCR plate. Data analysis for commercial qRT-PCR arrays was performed using the template provided online by SABiosciences. Normalized expression values for custom qRT-PCR data were determined using Microsoft Excel by first averaging the relative expression for each target gene (2-(Cqtarget gene-Cqreference gene)) across all biological replicates and then dividing the relative expression value of the experimental condition by the control condition (2-(ΔCqExperimental- ΔCqControl)). Error bars are derived from standard error of ΔCq values (Cqtarget gene-Cqreference gene) across all biological replicates. ACTB, GAPDH, and RPS18 were all used as reference genes. Normalized expression values for each target gene in all qRT-PCR experiments are derived from at least 3 biological replicates. A list of all custom qRT-PCR primers utilized is provided in Supplementary Tables 2 and 3.
Chromatin immunoprecipitation
Cells were treated with DMSO or TPA for 3 h at 37°C, 5% CO2 prior to execution of the ChIP protocol as described41. 1 μL of anti-Krt17 antibody or pre-immune sera was incubated per mg of total protein for all K17 ChIP assays. 10 μL of RFP-trap beads were used per IP condition for all mCherry-Aire ChIP assays. Antibody incubations occurred overnight at 4°C. The eluates were precipitated and resuspendend in 60 μL of sterile H2O with 1 μL used per PCR reaction (40 cycles) with SSO Advanced SYBR green mix (Bio-Rad) and primers as outlined in Supplemental Information. PCR products were separated by 1.5% agarose electrophoresis.
Electorphoretic mobility shift assays (EMSAs)
EMSAs were conducted using the EMSA kit (Promega) as previously described42 with modifications. A431 keratinocytes were serum-starved for 24-hrs followed by 1-hr treatment with TPA (200 nM) prior to isolation of nuclear extracts. For supershift analyses, nuclear extracts were incubated with 1 μL of either rabbit-IgG (Source), rabbit anti-p65 (Cell Signaling), pre-immune sera10, or anti-K1710 antibodies for 15 minutes at room temperature prior to incubation with 32P-labeled NF-κB oligonucleotide. For competition binding analyses, 50-fold excess non-labeled NF-κB or Oct1 (as a control) oligonucleotide was incubated for 15 minutes at room temperature with nuclear extract prior to incubation with 32P-labeled NF-κB oligonucleotide. Oligonucleotide labeling and gel shift assays were conducted following manufacturer's protocol (Promega, Gel Shift Assay System). Samples were resolved on a 6% DNA retardation gel (Invitrogen) in 0.5 × TBE buffer. Autoradiography was carried out on dried gels using phosphor-screens and phosphorimager.
Description of statistical methods
All error bars represent the standard deviation across biological replicates divided by the square root of the sample size (s.e.m.). Where technical replicates were conducted, these values were averaged to yield a single value per biological replicate. All p values were obtained by simple t-test with two-sided distribution and equal variance (Microsoft Excel), except as stated in Supplementary Figure 3.
Supplementary Material
1
2
Acknowledgments
The authors thank members of the Coulombe laboratory for support, Dr. Pärt Peterson (Aire constructs), Dr. Roland Foisner (subcellular fractionation), Ms. Monika Vladut-Talor (central tolerance study), and Ms. Janet Folmer (electron microscopy) for advice and assistance. These studies were supported by research grants CA160255 and AR44232 (to P.A.C.), GM111682 (to F.W.), and training grant CA009110 (to R.P.H.) all from the National Institutes of Health.
Footnotes
Author Contributions: R.P.H. and D.D. co-led the characterization of skin tumorigenesis in the HPV mouse model, with assistance from M.C.H. and A.B. R.P.H. led the studies performed in A431 cells and mouse keratinocytes in culture, with contributions from J.T.J., B.M.C., B.P., Y.G., and J.H. S.O. and D.C. conducted central tolerance study with assistance from R.P.H. and A.B. J.M.T. provided human BCC samples. W.Z. and F.W. performed the flow analyses. F.W. provided guidance to R.P.H. for EMSA assays. R.P.H., D.D. and P.A.C. contributed to experimental design and data analysis. R.P.H. and P.A.C. jointly wrote the manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 2.Karantza V. Keratins in health and cancer: more than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin L, Wang G. Keratin 17: a critical player in the pathogenesis of psoriasis. Med Res Rev. 2014;34:438–454. doi: 10.1002/med.21291. [DOI] [PubMed] [Google Scholar]
- 4.Chung BM, et al. Regulation of C-X-C chemokine gene expression by keratin 17 and hnRNP K in skin tumor keratinocytes. J Cell Biol. 2015;208:613–627. doi: 10.1083/jcb.201408026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Depianto D, Kerns ML, Dlugosz AA, Coulombe PA. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat Genet. 2010;42:910–914. doi: 10.1038/ng.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankar S, et al. A novel role for keratin 17 in coordinating oncogenic transformation and cellular adhesion in Ewing sarcoma. Mol Cell Biol. 2013;33:4448–4460. doi: 10.1128/MCB.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441:362–365. doi: 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- 8.Chen R, et al. Rac1 regulates skin tumors by regulation of keratin 17 through recruitment and interaction with CD11b+Gr1+ cells. Oncotarget. 2014;5:4406–4417. doi: 10.18632/oncotarget.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arbeit JM, Munger K, Howley PM, Hanahan D. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J Virol. 1994;68:4358–4368. doi: 10.1128/jvi.68.7.4358-4368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGowan KM, Coulombe PA. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J Cell Biol. 1998;143:469–486. doi: 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodworth CD, et al. Strain-dependent differences in malignant conversion of mouse skin tumors is an inherent property of the epidermal keratinocyte. Carcinogenesis. 2004;25:1771–1778. doi: 10.1093/carcin/bgh170. [DOI] [PubMed] [Google Scholar]
- 12.Mueller MM. Inflammation in epithelial skin tumours: old stories and new ideas. Eur J Cancer. 2006;42:735–744. doi: 10.1016/j.ejca.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Lee CH, Kim MS, Chung BM, Leahy DJ, Coulombe PA. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol. 2012;19:707–715. doi: 10.1038/nsmb.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong X, Coulombe PA. A novel mouse type I intermediate filament gene, keratin 17n (K17n), exhibits preferred expression in nail tissue. J Invest Dermatol. 2004;122:965–970. doi: 10.1111/j.0022-202X.2004.22422.x. [DOI] [PubMed] [Google Scholar]
- 15.Laan M, Peterson P. The Many Faces of Aire in Central Tolerance. Front Immunol. 2013;4:326. doi: 10.3389/fimmu.2013.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathis D, Benoist C. A decade of AIRE. Nat Rev Immunol. 2007;7:645–650. doi: 10.1038/nri2136. [DOI] [PubMed] [Google Scholar]
- 17.Metzger TC, Anderson MS. Control of central and peripheral tolerance by Aire. Immunol Rev. 2011;241:89–103. doi: 10.1111/j.1600-065X.2011.01008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kisand K, Peterson P. Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy: known and novel aspects of the syndrome. Ann N Y Acad Sci. 2011;1246:77–91. doi: 10.1111/j.1749-6632.2011.06308.x. [DOI] [PubMed] [Google Scholar]
- 19.Sillanpaa N, et al. Autoimmune regulator induced changes in the gene expression profile of human monocyte-dendritic cell-lineage. Mol Immunol. 2004;41:1185–1198. doi: 10.1016/j.molimm.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Giraud M, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A. 2012;109:535–540. doi: 10.1073/pnas.1119351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laan M, et al. Autoimmune regulator deficiency results in decreased expression of CCR4 and CCR7 ligands and in delayed migration of CD4+ thymocytes. J Immunol. 2009;183:7682–7691. doi: 10.4049/jimmunol.0804133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:6220. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 23.Kumar V, et al. The autoimmune regulator (AIRE), which is defective in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients, is expressed in human epidermal and follicular keratinocytes and associates with the intermediate filament protein cytokeratin 17. Am J Pathol. 2011;178:983–988. doi: 10.1016/j.ajpath.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 25.Barboro P, Ferrari N, Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352:152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Pitkanen J, Vahamurto P, Krohn K, Peterson P. Subcellular localization of the autoimmune regulator protein. characterization of nuclear targeting and transcriptional activation domain. J Biol Chem. 2001;276:19597–19602. doi: 10.1074/jbc.M008322200. [DOI] [PubMed] [Google Scholar]
- 27.Akiyoshi H, et al. Subcellular expression of autoimmune regulator is organized in a spatiotemporal manner. J Biol Chem. 2004;279:33984–33991. doi: 10.1074/jbc.M400702200. [DOI] [PubMed] [Google Scholar]
- 28.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaetani M, et al. AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome. Nucleic Acids Res. 2012;40:11756–11768. doi: 10.1093/nar/gks933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao J, Lowthert LA, Ghori N, Omary MB. The 70-kDa heat shock proteins associate with glandular intermediate filaments in an ATP-dependent manner. J Biol Chem. 1995;270:915–922. doi: 10.1074/jbc.270.2.915. [DOI] [PubMed] [Google Scholar]
- 31.Kumeta M, Hirai Y, Yoshimura SH, Horigome T, Takeyasu K. Antibody-based analysis reveals “filamentous vs. non-filamentous” and “cytoplasmic vs. nuclear” crosstalk of cytoskeletal proteins. Exp Cell Res. 2013;319:3226–3237. doi: 10.1016/j.yexcr.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell. 2010;140:123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 34.Lenardo MJ, Baltimore D. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 35.Pasparakis M. Role of NF-kappaB in epithelial biology. Immunol Rev. 2012;246:346–358. doi: 10.1111/j.1600-065X.2012.01109.x. [DOI] [PubMed] [Google Scholar]
Methods Only References
- 36.McGowan KM, et al. Keratin 17 null mice exhibit age- and strain-dependent alopecia. Genes Dev. 2002;16:1412–1422. doi: 10.1101/gad.979502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichelt J, Haase I. Establishment of spontaneously immortalized keratinocyte lines from wild-type and mutant mice. Methods Mol Biol. 2010;585:59–69. doi: 10.1007/978-1-60761-380-0_5. [DOI] [PubMed] [Google Scholar]
- 38.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lessard JC, et al. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc Natl Acad Sci U S A. 2013;110:19537–19542. doi: 10.1073/pnas.1309576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adamson KA, Pearce SH, Lamb JR, Seckl JR, Howie SE. A comparative study of mRNA and protein expression of the autoimmune regulator gene (Aire) in embryonic and adult murine tissues. J Pathol. 2004;202:180–187. doi: 10.1002/path.1493. [DOI] [PubMed] [Google Scholar]
- 41.Wan F, et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Fu K, et al. Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells. Nat Commun. 2013;4:1909. doi: 10.1038/ncomss2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2