5′ UTR m6A Promotes Cap-Independent Translation (original) (raw)
. Author manuscript; available in PMC: 2016 Nov 5.
SUMMARY
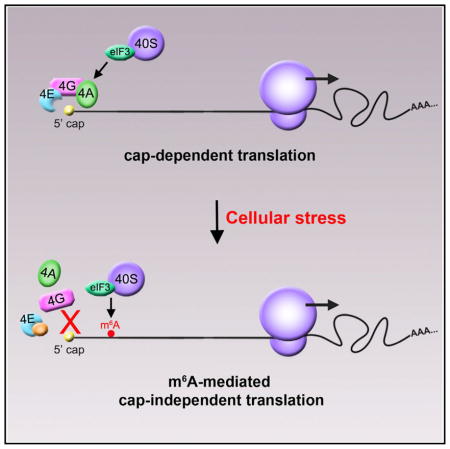
Protein translation typically begins with the recruitment of the 43S ribosomal complex to the 5′ cap of mRNAs by a cap-binding complex. However, some transcripts are translated in a cap-independent manner through poorly understood mechanisms. Here, we show that mRNAs containing _N_6-methyladenosine (m6A) in their 5′ UTR can be translated in a cap-independent manner. A single 5′ UTR m6A directly binds eukaryotic initiation factor 3 (eIF3), which is sufficient to recruit the 43S complex to initiate translation in the absence of the cap-binding factor eIF4E. Inhibition of adenosine methylation selectively reduces translation of mRNAs containing 5′UTR m6A. Additionally, increased m6A levels in the Hsp70 mRNA regulate its cap-independent translation following heat shock. Notably, we find that diverse cellular stresses induce a transcriptome-wide redistribution of m6A, resulting in increased numbers of mRNAs with 5′ UTR m6A. These data show that 5′ UTR m6A bypasses 5′ cap-binding proteins to promote translation under stresses.
Graphical Abstract
INTRODUCTION
For most cellular mRNAs, the first step of mRNA translation involves recognition of the 5′ 7-methylguanosine (m7G) cap by eukaryotic initiation factor 4E (eIF4E), which is a subunit of the heterotrimeric eIF4F complex. 5′ cap-bound eIF4F then recruits the small (40S) ribosomal subunit associated with various translation initiation factors, enabling efficient translation of eukaryotic mRNAs.
However, some mRNAs are translated in a cap-independent manner. These capped mRNAs do not require eIF4E and are translated under basal cellular conditions, as well as conditions in which eIF4E activity is compromised, such as cellular stress states, viral infection, and diseases such as cancer (Stoneley and Willis, 2004). Although viral mRNAs can exhibit cap-independent translation due to the presence of highly structured internal ribosome entry site (IRES) motifs in the 5′ UTR, correspondingly complex structures are rarely found in eukaryotic mRNAs undergoing cap-independent translation (Stoneley and Willis, 2004). Thus, the mechanism of cap-independent translation in cellular mRNAs remains poorly understood.
A feature of many eukaryotic mRNAs is _N_6-methyladenosine (m6A), a reversible base modification seen in the 3′ UTR, coding sequence, and 5′ UTR (Dominissini et al., 2012; Meyer et al., 2012). Although the function of m6A in 3′UTRs has been explored (Wang et al., 2014a, 2014b, 2015), the function of m6A in 5′ UTRs remains unknown. Here, we show that m6A in the 5′ UTR functions as an alternative to the 5′ cap to stimulate mRNA translation. Using both in vitro reconstitution approaches and translation assays in cellular lysates deficient in eIF4E activity, we define a unique translation initiation mechanism that does not require the 5′ cap. We show that the m6A in the 5′ UTR can bind eukaryotic initiation factor 3 (eIF3). Transcriptome-wide ribosome profiling analysis indicates that the translation of 5′ UTR m6A-containing mRNAs is reduced upon depletion of the m6A methyltransferase, METTL3, while mRNAs containing m6A elsewhere within the transcript fail to show this effect. The importance of 5′ UTR m6A residues for cellular mRNA translation is demonstrated by both ribosome profiling analysis and detection of changes to global m6A distribution in 5′UTRs in response to cellular stress. Thus, 5′ UTR m6A residues are linked to cellular stress states and provide a mechanism to bypass the m7G cap requirement for mRNA translation, enabling a cap-independent mode of translation initiation.
RESULTS
Ribosomal Initiation Complexes Assemble on m6A-Containing mRNAs Independently of the Cap-Binding Protein eIF4E
Although m6A is predominantly localized near stop codons and in 3′ UTRs in several thousand mRNAs, hundreds of cellular mRNAs contain m6A within their 5′ UTR (Linder et al., 2015; Meyer et al., 2012), and the function of these m6A residues is unknown. Since the 5′ UTR is important in regulating translation initiation, we considered the possibility that 5′ UTR-localized m6As might influence this process. On most eukaryotic mRNAs, translation begins with assembly of a 43S preinitiation complex, comprising a 40S ribosomal subunit, a eukaryotic initiation factor 2 (eIF2)-GTP/Met-tRNAiMet ternary complex, and eIFs 3, 1, and 1A (Jackson et al., 2010). 43S complexes are typically recruited to mRNA by a cap-binding complex, eIF4F. eIF4F consists of three subunits: eIF4E, which binds the m7G 5′ cap; eIF4A, an RNA helicase; and eIF4G, a scaffold that also binds eIF3, thereby recruiting the 43S complex. After attachment, 43S complexes scan to the initiation codon, where they form 48S initiation complexes (Jackson et al., 2010).
To investigate the effect of m6A on translation initiation, we used toeprinting, an approach for reconstituting assembly of 48S complexes on mRNA. In toeprinting, ribosomal complexes are assembled on mRNA 5′ UTRs using purified translational components (40S subunits, initiation factors and Met-tRNAiMet) (Pestova and Kolupaeva, 2002). Formation of the 48S complex at the start codon is then monitored by reverse transcriptase-mediated extension of a [32P]-labeled primer annealed to ribosome-bound mRNA. cDNA synthesis is arrested by the 40S ribosome subunit, yielding characteristic toeprints at its leading edge, +15–17 nt downstream of the initiation codon. This assay can identify the initiation factors and sequence features of 5′ UTRs that are required for initiation and has been used in mechanistic studies of viral IRESs (Pestova and Hellen, 2003).
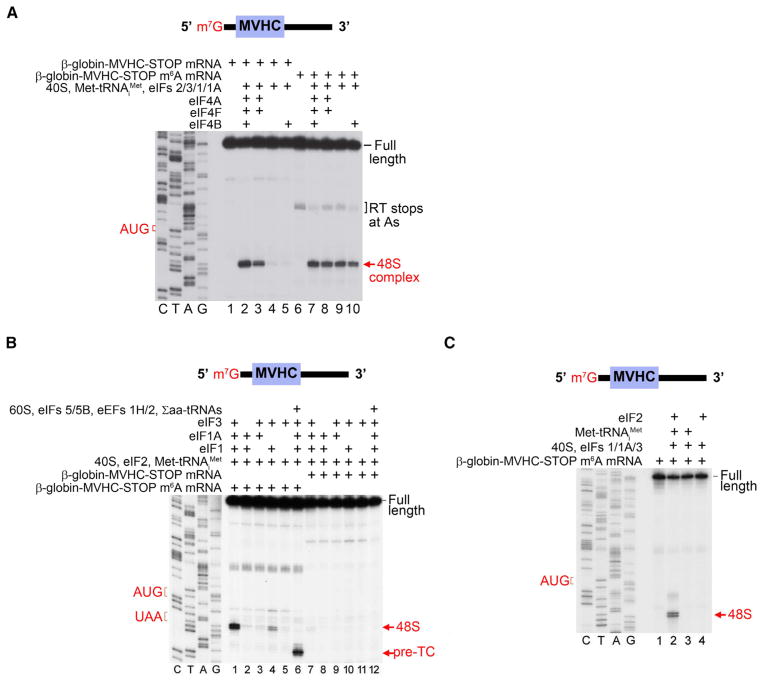
To test the role of m6A in 48S complex formation, we performed toeprinting with 5′-capped mRNAs comprising the 54-nt-long β-globin 5′ UTR followed by a short coding sequence, stop codon, and 3′ UTR. Consistent with previous studies (Pestova and Kolupaeva, 2002), 48S initiation complexes were detected at the start codon of A-containing mRNA in the presence of the complete set of eIFs (1, 1A, 2, 3, 4A, 4B, and 4F), and omission of group 4 eIFs nearly abrogated 48S complex formation (Figure 1A, compare lanes 2 and 4). This is consistent with the known role for the eIF4 cap-binding complex in recruiting the 43S complex to mRNA (Gingras et al., 1999).
Figure 1. 5′UTR m6A Enables Ribosome Binding to mRNA in the Absence of Cap-Binding Proteins.
(A) 5′ UTR methylation permits 48S initiation complex formation in the absence of the group 4 eIFs. In vitro transcribed, capped mRNAs encoding a MVHC tetrapeptide and containing either A or m6A were incubated with purified mammalian translation initiation components. Subsequent toeprinting analysis using a radiolabeled primer then revealed whether 48S initiation complexes were formed. Positions of the initiation codon, full-length cDNA, and the 48S complex are shown on the sides of the panel. Lanes C/T/A/G depict the corresponding DNA sequence. When unmethylated mRNA is used (lanes 1–5), 48S complexes are only formed when the cap-binding complex eIF4F is present (lanes 2 and 3). When eIF4F is absent, 48S complex formation on unmethylated mRNA is impaired (lanes 4 and 5). However, when mRNA with m6A in the 5′ UTR is used, 48S complex formation is observed even in the absence of eIF4F (lanes 9 and 10; compare to lanes 7 and 8 where eIF4F is present).
(B) eIFs1, 1A, and 3 are required for efficient m6A-induced cap-independent 48S complex formation. Toeprinting assays were performed as in (A) using A- or m6A-containing mRNAs and in the presence of various translation initiation components as indicated. m6A-containing mRNA exhibits robust 48S complex assembly in the absence of eIF4F, whereas A-containing mRNA does not (compare lanes 1 and 7). Efficient m6A-mediated 48S complex assembly is also dependent on the presence of eIFs1 and 1A, which is consistent with the known roles of these proteins in promoting scanning and AUG recognition (compare lanes 1 with lanes 2, 4, and 5). Removal of eIF3 also abolishes 48S complex assembly on m6A-containing mRNA (compare lanes 1 and 2), indicating that eIF3 is required for m6A-mediated 48S complex formation. Addition of 60S subunits, eIF5, eIF5B, eEF1H, eEF2, and aa-tRNAs resulted in the appearance of toeprints corresponding to pre-termination complexes at the stop codon, indicating that m6A-recruited 48S complexes are fully functional (lane 6).
(C) Omission of eIF2 from toeprinting assays results in the absence of 48S complexes (compare lanes 2 and 3), indicating that eIF2 is required for 48S complex assembly on m6A-containing mRNA.
When we used mRNAs that were in vitro transcribed to contain m6A, we found that 48S complexes readily assembled after addition of the complete set of eIFs, as was seen with unmethylated mRNA. However, unlike the unmethylated mRNA, 48S complexes formed on m6A-containing mRNA even in the absence of group 4 eIFs (Figure 1A). Thus, initiation on m6A-containing mRNA is distinct from initiation on mRNA lacking m6A and does not require the eIF4 cap-binding complex.
To further establish the factor requirements for initiation on m6A-containing mRNA, we selectively omitted each initiation factor and performed toeprinting. These experiments show that efficient initiation on m6A-containing mRNA only requires the presence of eIFs1, 1A, 2, 3, and the 40S subunit (Figures 1B and 1C). 48S complexes that formed on m6A-containing mRNA in the absence of group 4 eIFs were functional, as addition of the 60S ribosomal subunit, Σaa-tRNAs, and factors required for subunit joining and elongation (eIF5, eIF5B, eEF2, and eEF1H) resulted in formation of 80S ribosomes that underwent efficient elongation and yielded pre-termination complexes at the stop codon (Figure 1B). Thus, translation-competent 48S complexes can form on m6A-containing mRNA in the absence of eIF4E.
m6A Enables Translation in a 5′ Cap-Independent Manner in Cell-Free Extracts
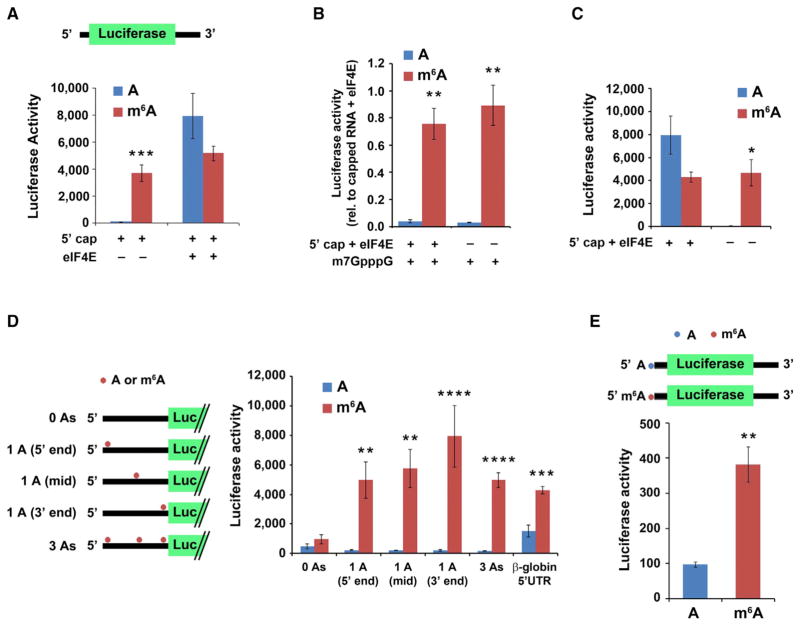
We next asked if m6A induces eIF4E-independent translation in cell-free extracts. To investigate this, we used a HeLa extract that has low eIF4E activity (Mikami et al., 2006) (Figures S1A and S1B) and thus provides an ideal system for studying eIF4E-independent translation. Indeed, addition of a capped, nonmethylated luciferase-encoding mRNA containing the β-globin 5′ UTR to the HeLa extract did not produce measureable luciferase activity unless eIF4E was added (Figure 2A). Thus, cap-dependent translation in this extract is dependent on exogenous eIF4E.
Figure 2. m6A within the 5′ UTR Enables Cap-Independent Translation of mRNA.
(A) 5′ UTR m6A permits mRNA translation without the need for the cap-binding protein eIF4E. In vitro translation was performed using a HeLa cell extract mixed with luciferase-encoding, capped mRNA containing either A or m6A. Protein production was measured by quantifying luciferase activity. Cap-dependent translation is observed from both methylated and unmethylated mRNAs in the presence of eIF4E. However, when eIF4E is absent, only the m6A-containing mRNA is translated (n = 4; mean ± SD; ***p < 0.0001).
(B) Presence of a 5′ cap analog is unable to abolish m6A-induced mRNA translation. Luciferase mRNAs were translated as in (A). 1 mM free cap analog (m7GpppG) was added to sequester cap-binding proteins. Addition of m7GpppG abolishes cap-dependent translation of unmethylated mRNA (left) but is unable to abolish the cap-independent translation induced by m6A (right). Levels of luciferase activity are shown relative to capped mRNA + 10 pmole eIF4E (n = 3; mean ± SD; *p < 0.01, **p < 0.001).
(C) In vitro translation was performed using luciferase-encoding mRNA containing A or 50% m6A and with or without a 5′cap as indicated. While unmethylated, capped mRNA + 10 pmole eIF4E is robustly translated, the unmethylated, uncapped mRNA fails to be translated. However, m6A-containing mRNA is efficiently translated even when no 5′ cap is present (n = 3; mean ± SD; *p < 0.01).
(D) m6A residues in the coding sequence do not induce cap-independent translation. Uncapped, luciferase-encoding mRNAs containing either the natural β-globin 5′ UTR or a modified β-globin 5′ UTR containing either zero, one, or three A residues as indicated were used for in vitro translation assays. Translation of m6A-containing mRNA with zero A residues in the 5′ UTR was markedly diminished, indicating that coding sequence m6A residues are unable to induce cap-independent translation. However, when a single m6A was added to the 5′ UTR, the transcripts were robustly translated. Methylated 5′ UTRs with a single A near the 5′ end, the middle (mid), or near the 3′ end all showed similar levels of translation (n = 3; mean ± SD; **p < 0.001, ***p < 0.0001, ****p < 0.00001). Schematic shows the distribution of A residues within each β-globin 5′ UTR variant (the unmodified β-globin 5′ UTR contains 17 A residues).
(E) mRNA with a single m6A within the 5′ UTR and no m6As in the remainder of the transcript induces cap-independent translation. Uncapped, luciferase-encoding mRNAs, which contained either a single adenosine 5′-monophosphate (AMP) or _N_6-methyladenosine 5′-monophosphate (m6AMP) at the 5′ end, were used for in vitro translation. Only the m6A-containing mRNA was translated, demonstrating that a single 5′ end m6A residue is capable of inducing cap-independent translation (n = 3; mean ± SD; **p < 0.001). The reduced translation efficiency of this mRNA compared to mRNAs with internally methylated 5′ UTRs is likely due to inefficient incorporation of m6A residues at the 5′ end by T7 RNA polymerase.
See also Figure S1.
We next used HeLa extracts to determine if transcripts containing m6A require eIF4E. In contrast to the mRNA containing exclusively A, 5′-capped mRNA containing 50% m6A was readily translated even inthe absence of added eIF4E (Figure 2A). Furthermore, addition of 1 mM m7GpppG, a cap analog that sequesters cap-binding proteins (Ray et al., 2006), abolished translation of 5′-capped, A-containing mRNA but had no effect on m6A-containing mRNA (Figure 2B). Lastly, A-containing mRNA synthesized without a cap was not translated, whereas m6A-containing, uncapped mRNA was readily translated (Figure 2C). The increased translation of m6A-containing mRNA in these experiments was not due to increased stability of m6A-containing mRNA, as RT-qPCR and radiolabeled mRNA stability measurements indicated similar levels of A- and m6A-containing luciferase mRNA after incubation with HeLa extracts (Figures S1C–S1F). Collectively, these data indicate that translation of m6A-containing mRNA exhibits marked independence of the 5′ cap and eIF4E.
A Single m6A Is Sufficient to Induce Cap-Independent Translation
Since the mRNAs used in the in vitro translation assays have m6A throughout the transcript, it is unclear if the translational effects are due to m6A in the 5′ UTR or elsewhere in the mRNA. To determine the contributions of specific m6A residues to cap-independent translation, we examined mRNAs that only contain m6A in the coding sequence. Uncapped, luciferase-encoding mRNAs that contained zero m6A residues within the 5′ UTR showed no translation, indicating that m6A residues in the coding sequence are unable to induce cap-independent translation (Figure 2D). However, addition of a single m6A residue at the beginning, middle, or end of the 5′ UTR was sufficient to markedly induce cap-independent translation (Figure 2D).
To determine if a single 5′ UTR m6A residue can promote cap-independent translation, we used uncapped luciferase-encoding mRNAs that contain m6A as the first transcribed nucleotide (Supplemental Experimental Procedures). This mRNA contains a single m6A residue in the 5′ UTR, and the remainder of the As within the transcript are unmethylated. For mRNAs lacking m6A, negligible luciferase synthesis was detected (Figure 2E). However, transcripts containing a single 5′ m6A were readily translated (Figure 2E). Notably, the level of translation induced by a 5′ m6A is less than the translation induced by a single m6A residue located internally within the 5′ UTR, which likely reflects inefficient incorporation of m6A at the first position of 5′ m6A-containing transcripts (see Experimental Procedures). Collectively, these experiments indicate that a single m6A can induce cap-independent translation.
To determine whether m6A-mediated cap-independent translation is a specific effect caused by the presence of m6A, we synthesized uncapped luciferase transcripts containing A, m6A, or other modified nucleotides, such as _N_1-methyladenosine, 2′-_O_-methyladenosine, pseudouridine, and 5-methylcytosine. In each case, there was negligible luciferase synthesis unless m6A was present (Figure S1G).
We next asked if the effect of m6A reflects impaired base pairing caused by modification of the _N_6 position (Roost et al., 2015). However, mRNA containing _N_6-propargyladenosine, which contains a slightly larger modification compared to a methyl group at the _N_6 position, failed to undergo cap-independent translation (Figure S1G). Thus, m6A-induced structural changes are unlikely to account for the cap independence conferred by m6A.
m6A-Induced Translation Initiation Occurs through a 5′ End-Dependent Mechanism
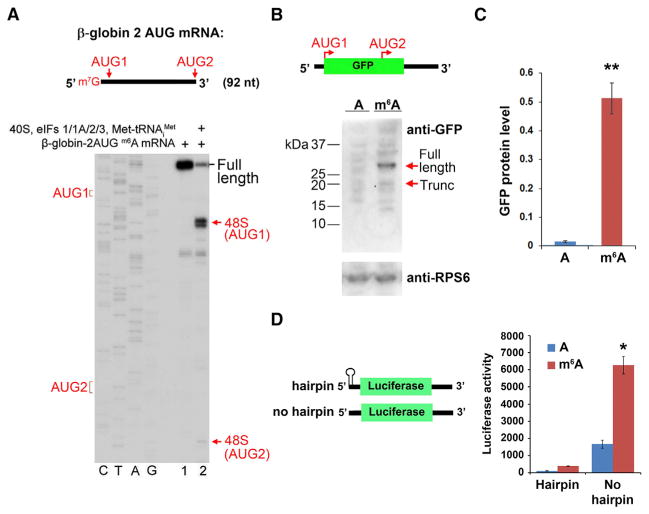
Our results indicate that m6A residues within the 5′ UTR are capable of promoting cap-independent translation. However, the majority of m6A residues are found in the coding sequence and 3′ UTR (Meyer et al., 2012). We therefore asked if these internal m6A residues can induce internal ribosome entry. To test this, we synthesized an m6A-containing β-globin mRNA in which the wild-type AUG initiation codon was removed and two new AUG triplets were introduced upstream and downstream of the native position (Figure 3A). When this mRNA was incubated with 40S, eIFs1/1A/2/3, and Met-tRNAiMet, 48S complexes occurred almost exclusively at the first AUG, with very low levels of detectable 48S complex formation at the downstream AUG (Figure 3A). These data suggest that m6A preferentially induces translation at the first suitable start codon in the mRNA as opposed to promoting translation through an internal entry-based mechanism.
Figure 3. m6A-Mediated Translation Occurs through a 5′ End-Dependent Mechanism.
(A) Toeprinting assays were performed using a capped, m6A-containing mRNA containing the β-globin 5′ UTR sequence, which was modified to include two AUG initiation codons (“AUG1” and “AUG2” in the schematic). The majority of 48S complexes were assembled at AUG1, with negligible levels of 48S complexes detected at AUG2.
(B) Uncapped, A-, or m6A-containing mRNAs encoding GFP were used for in vitro translation. The mRNA contains two near-kozak start codons: AUG 1 encodes the full-length GFP protein, and internally localized AUG2 encodes an in-frame truncated (~17 kDa) protein comprising the C-terminal portion of GFP. Full-length and truncated GFP protein levels (sizes indicated by arrows) were measured by western blot. m6A primarily promotes translation of the full-length protein and fails to induce internal entry-mediated translation from AUG2. Levels of the ribosomal protein RPS6 are shown as a loading control.
(C) Quantification of full-length GFP protein levels in (B) shows increased protein expression of methylated mRNA versus unmethylated mRNA (n = 3; mean ± SD; **p < 0.001).
(D) The presence of a stable hairpin at the beginning of the 5′ UTR to block 5′ end entry severely attenuates m6A-mediated translation (n = 3; mean ± SD; *p < 0.01).
See also Figure S1.
Next, we used HeLa cell lysates to in vitro translate a GFP reporter mRNA containing an internal near-Kozak AUG in addition to the natural AUG encoding full-length GFP. However, we failed to observe m6A-mediated translation of the ~17 kDa product produced from the internal AUG and instead observed robust translation of the full-length protein produced from the first AUG (Figures 3B, 3C, and S1H). These results are consistent with the toeprinting experiments and suggest that m6A preferentially induces translation at the first acceptable start codon.
The selective use of the first AUG for translation initiation suggests a model of m6A-mediated initiation that involves a 5′ end-dependent scanning mechanism as opposed to internal ribosomal entry. A similar mode of initiation, which is also cap independent but shows 5′-end dependence, was recently described for mRNA containing in its 5′ UTR an eIF4G-binding viral IRES-domain (Terenin et al., 2013). Additionally, cap-independent, 5′ end-dependent mechanisms of translation initiation have previously been observed in assays using rabbit reticulocyte lysates (De Gregorio et al., 1998). To test directly whether m6A promotes entry through the 5′ end, we used an uncapped, luciferase-encoding mRNA that contains a stable hairpin at the extreme 5′ end of the mRNA to block 5′ end-dependent ribosome entry. We found that the presence of this hairpin markedly reduced the robust translation of m6A-containing mRNA that is normally observed (Figure 3D). Thus, m6A-mediated initiation requires an accessible 5′-terminal end on the mRNA. Taken together, these data indicate that 5′ UTR m6As are distinct from classical viral IRES elements since m6A promotes recruitment of ribosomal preinitiation complexes to the 5′ end of mRNA, rather than enabling internal ribosome entry.
eIF3 Selectively Binds m6A-Containing RNA
We next asked how m6A is recognized to induce translation of mRNAs. The in vitro 48S reconstitution assays showed that recruitment of the 43S preinitiation complex to m6A-containing mRNA only requires eIFs 1, 1A, 2, and 3 and the 40S subunit. Thus, one of these components binds m6A.
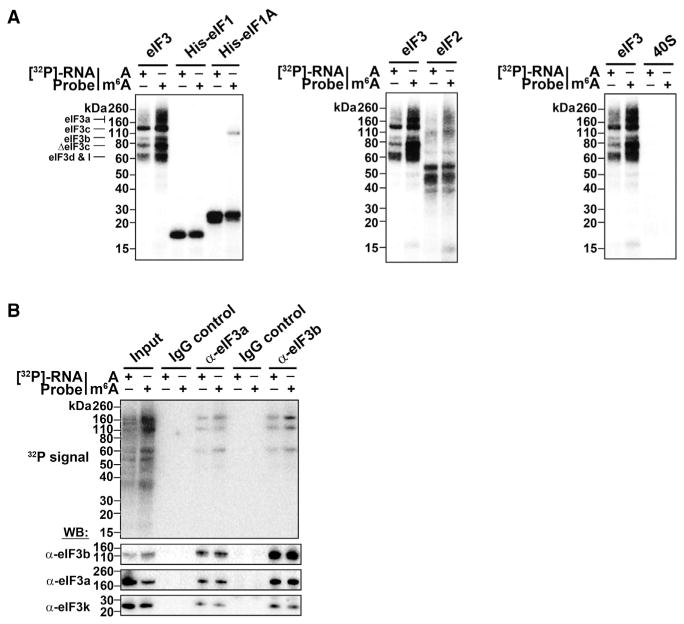
To test which of these factors interacts with m6A, we used an m6A crosslinking assay in which a [32P]-labeled RNA probe containing a single A or m6A in its naturally occurring GAC context was UV-crosslinked to each translational component. Crosslinked proteins were then detected by SDS-PAGE and autoradiography.
eIFs 1, 1A, and 2 and the 40S subunit showed equal levels of crosslinking to the A- and m6A-containing probes (Figures 4A and S2A). However, crosslinking of eIF3 to the m6A-containing probe was substantially increased compared to the A-containing probe, suggesting that this factor constitutes the major m6A-binding activity of the 43S complex (Figures 4A, S2B, and S2C).
Figure 4. The 43S Complex Component eIF3 Binds m6A.
(A) Indicated proteins/protein complexes were incubated with radiolabeled A- or m6A-containing RNA probes and crosslinked. Unbound RNAs were then removed with RNase I, proteins were separated by SDS-PAGE, and radioactively-labeled RNAs were detected. eIF1, eIF1A, eIF2, and the 40S ribosomal subunit show no preferential crosslinking to methylated RNA. However, eIF3 preparations exhibit strong crosslinking to methylated RNA at bands around 60 kD, 80 kD, and 110–160 kD, which correspond to multiple subunits of the eIF3 complex as indicated.
(B) Crosslinking assays were performed as in (A) using the HeLa cell extracts utilized in in vitro translation assays. The eIF3 complex was immunoprecipitated using antibodies against eIF3a or eIF3b, and proteins containing crosslinked RNA were detected. Both eIF3 antibodies precipitated proteins that preferentially crosslinked to m6A RNA. Immunoprecipitation using rabbit and mouse IgG control antibodies are shown as negative controls. Western blotting for the indicated proteins indicates their enrichment following immunoprecipitation (bottom). The input lanes throughout have 25% of the material loaded for the IP lanes.
See also Figures S2 and S3.
The preferential binding of eIF3 to m6A was not affected by changing the position of the m6A together with its context nucleotides within the probe (Figure S2D). However, when the natural nucleotide context of m6A was changed from GAC to UAC or CAG, the m6A-containing probe showed significantly reduced crosslinking to eIF3 (Figure S2E). Thus, efficient eIF3 crosslinking to m6A-containing RNA occurs when the probe contains m6A within its natural sequence context. Furthermore, when we subjected mRNAs that contained a single m6A residue within their 5′ UTR to in vitro translation, we found that m6A residues in a GAC context promoted robust cap-independent translation, whereas m6As in a UAC or CAG exhibited markedly reduced translation (Figure S2F). These data indicate that eIF3 preferentially binds to m6A residues in their natural sequence context to promote cap-independent translation.
eIF3 is a large multiprotein complex comprising 13 subunits (a–m) (des Georges et al., 2015) that interacts with mRNA in 48S complexes (Pisarev et al., 2008). UV-crosslinking studies showed that the interaction between eIF3 and RNA occurs at a multisubunit interface (Lee et al., 2015). Similarly, in our cross-linking assays, the m6A-containing probe induced strong labeling of several protein bands, ranging in molecular weight from ~60 to ~160 kDa (Figures 4A and S2A–S2E). Particularly strong labeling was observed in the area of ΔeIF3a/eIF3c, ΔeIF3c, and eIF3d/eIF3I (Figures S2A–S2E). These data suggest that m6A-containing RNA may interact with a multisubunit interface within eIF3.
To further explore the binding of m6A-containing RNA to eIF3, we used HeLa cell lysates. Crosslinking using a radioactive m6A-containing RNA probe resulted in the labeling of specific protein bands that were increased relative to the A-containing probe (Figure 4B). Immunoprecipitation of crosslinked extracts using either of two eIF3 subunit-specific antibodies selectively precipitated these bands, confirming that the increased binding to m6A-containing RNA was mediated by eIF3. Immunoprecipitation with a control antibody recognizing a different initiation-factor-associated protein (ABCF1) did not precipitate these bands (Figures 4B, S3A, and S3B). Thus, these data further suggest that m6A-containing RNA interacts with eIF3.
The m6A-binding protein YTHDF1 interacts with a diverse set of proteins, including eIF3 (Wang et al., 2015). Thus, we considered the possibility that recruitment of eIF3 to m6A-containing RNA in the in vitro translation and crosslinking assays is mediated by a YTH-family m6A-binding protein. However, silver staining of all the initiation factors used in the toeprinting assays failed to show protein bands in the ~60–64 kD range of these proteins (Figure S2). Additionally, mass spectrometry analysis of the purified eIF3 did not reveal YTH family proteins (Figure S3C) (des Georges et al., 2015). Finally, YTHDF1 was not present in the highly purified eIF3 preparations used in our crosslinking assays, nor were any of the related YTH-domain containing family of m6A binding proteins (Figure S3D) (des Georges et al., 2015). Thus, these data support the idea that eIF3 is able to directly bind m6A.
To determine whether eIF3 binds m6A in cells, we performed PAR-iCLIP to identify zero-distance binding sites of eIF3 in cellular mRNAs. eIF3a-binding sites were primarily localized to 5′ UTRs of mRNAs and showed a high degree of overlap with eIF3-binding sites reported previously (Lee et al., 2015) (Figures S4A and S4B).
To determine whether eIF3a binds to sites of m6A in 5′ UTRs, we evaluated the overlap of eIF3a-binding sites with m6A residues mapped at single-nucleotide-resolution in 5′ UTRs (Linder et al., 2015). To test this, we used a permutation-based approach in which eIF3a-binding sites were randomized while preserving the distribution and positional bias of eIF3a PAR-iCLIP tags in 5′ UTRs. Multiple permutations (n > 100) were used, and the statistical significance of overlap between eIF3 PAR-iCLIP sites and m6A residues was evaluated (Supplemental Experimental Procedures). We found a statistically significant overlap between m6A residues and eIF3-binding sites in 5′ UTRs, with 35% of 5′ UTR m6A residues overlapping with eIF3 sites (Figures S4C–S4E). Since single-nucleotide-resolution m6A mapping distinguishes between m6A residues and the m6Am residues that exist as part of the 5′ cap in some mRNAs (Kruse et al., 2011; Linder et al., 2015), we were able to determine that this overlap was specific to m6A residues within 5′ UTRs (Figures S4C and S5A). Taken together, these results support the idea that eIF3 is associated with m6A residues in the 5′ UTRs of cellular mRNAs.
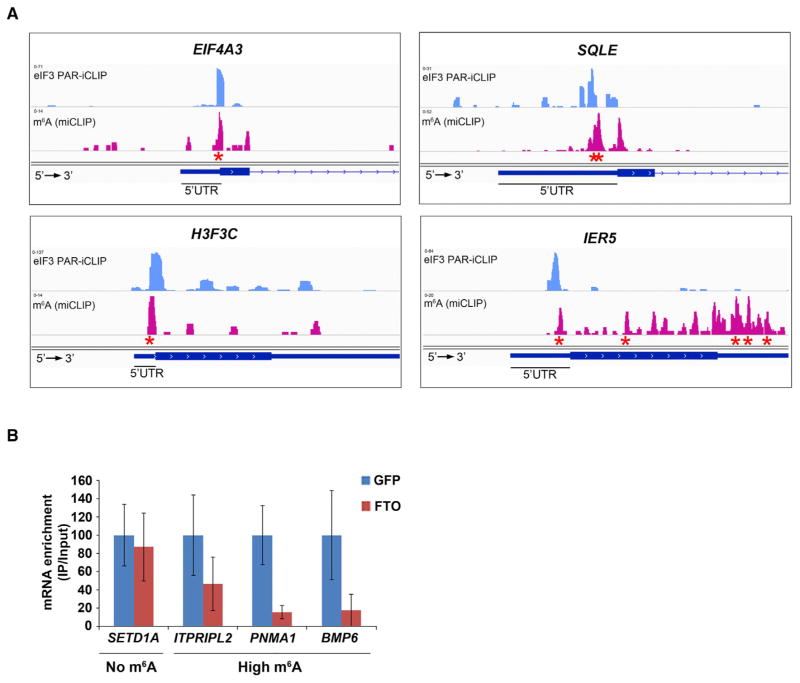
To further test the physiological association of eIF3 and m6A predicted by the PAR-iCLIP analysis, we performed eIF3 protein/RNA immunoprecipitation from HEK293 cells expressing the m6A-demethylating enzyme (Jia et al., 2011), Fto. The abundance of target mRNA 5′ UTRs in the eIF3-bound fraction was then measured using RT-qPCR with primers that amplify the 5′ UTR regions containing the m6A residue. mRNAs that contain a high stoichiometry m6A site within their 5′ UTR (Meyer et al., 2012) were substantially depleted in the eIF3-bound fraction following Fto overexpression (Figure 5B). In contrast, eIF3 immunoprecipitation of a control mRNA deficient in 5′ UTR m6A (Meyer et al., 2012) was unaffected by Fto overexpression (Figure 5B). Taken together, these data support the idea that eIF3 interacts with mRNAs in an m6A-dependent manner in cells.
Figure 5. eIF3 Binding Sites within Cellular mRNAs Localize to Sites of m6A Residues within the 5′ UTR.
(A) Shown are read clusters from both eIF3 PAR-iCLIP (light blue) and single-nucleotide-resolution m6A mapping (Linder et al., 2015) (miCLIP; red) for four representative mRNAs (EIF4A3, H3F3C, SQLE, and IER5). eIF3a PAR-iCLIP read clusters exhibit highly specific overlap with m6A mapping clusters at internal positions within 5′ UTRs. This co-localization is specific to 5′ UTRs, as mRNAs that contain multiple m6A residues in the CDS or 3′ UTR fail to show eIF3a binding at these sites (exemplified by IER5). Red asterisks indicate the location of individual m6A sites identified at single-nucleotide resolution.
(B) eIF3 binds to the 5′ UTR of cellular mRNAs in an m6A-dependent manner. HEK293 cells were transfected with GFP- or Fto-overexpression plasmids, and eIF3 immunoprecipitation was performed to isolate eIF3-bound mRNAs. Bound mRNAs were quantified by RT-qPCR using 5′ UTR-specific primers. 5′ UTRs of mRNAs that contain high levels of m6A exhibited reduced binding to eIF3 after overexpression of Fto. 5′ UTRs that do not contain m6A exhibited no change in eIF3 binding following Fto overexpression (n = 3; mean ± SEM).
See also Figures S4 and S5.
m6A within the 5′ UTR Promotes Translation of Cellular mRNAs
To address whether mRNAs that contain 5′ UTR m6A residues possess enhanced translation in cells, we examined ribosome profiling-based measurements of mRNA translation efficiency (TE) in HeLa cells depleted of the m6A methyltransferase enzyme, METTL3, which results in depletion of all m6A residues in cells (Wang et al., 2015). We examined the TE of mRNAs based on the location of their m6A residues identified by single-nucleotide-resolution m6A mapping (Linder et al., 2015). Compared to mRNAs that lack m6A, we found that transcripts that contain m6A residues within the coding sequence or 3′ UTR show no significant change in TE in METTL3-depleted cells (Figures 6A and 6B). Similarly, mRNAs that contain m6A residues near the stop codon do not show reduced translation in METTL3-depleted cells. However, mRNAs containing 5′ UTR m6A residues showed a large reduction in TE following METTL3 depletion, suggesting a preferential role for 5′ UTR m6A in promoting mRNA translation (Figures 6A, 6B, and S5B). Residual translation may reflect ongoing cap-dependent translation in METTL3-deficient cells. The translation of mRNAs containing 5′ UTR m6A residues was not suppressed in cells depleted of YTHDF1 (Figure S5B), which is consistent with the idea that 5′ UTR m6A promotes translation through eIF3. Taken together, these data suggest that m6A residues in the 5′ UTR enhance the translation of mRNAs in cells.
Figure 6. m6A Mediates Stress-Induced Translation of Hsp70.
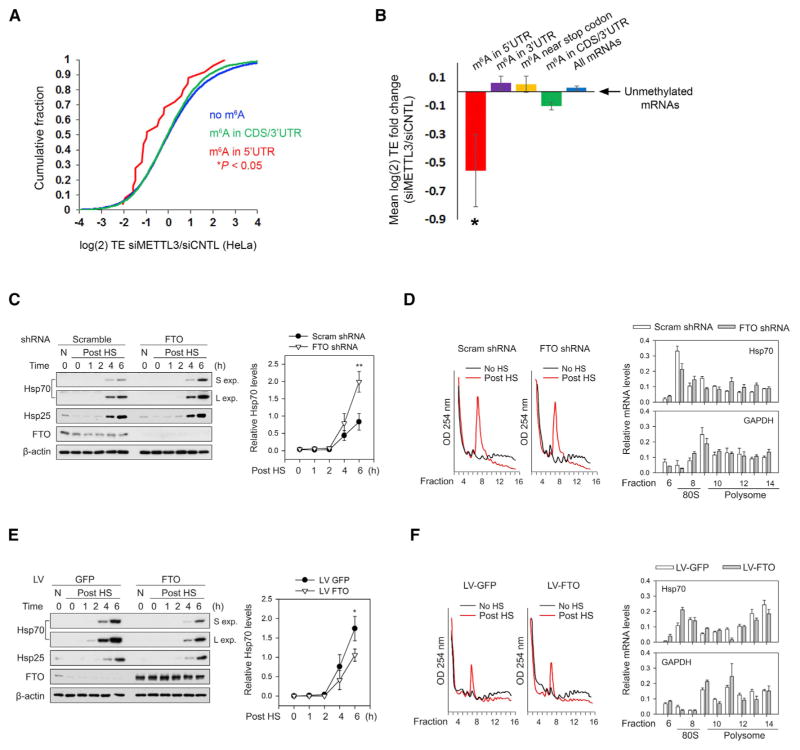
(A) Depletion of the m6A methyltransferase, METTL3, decreases the TE of mRNAs with 5′ UTR m6A. Ribosome profiling data from HeLa cells expressing METTL3 or control siRNAs (Wang et al., 2015) were used to determine changes in TE for various classes of mRNAs defined by single-nucleotide-resolution m6A mapping. Compared to nonmethylated mRNAs (blue), transcripts with m6A residues in the coding sequence (CDS) or 3′ UTR (green) exhibit only a marginal decrease in TE. However, mRNAs containing m6A within the 5′ UTR (red) show a large reduction in TE. p values were calculated using the Mann-Whitney test.
(B) TEs of various classes of m6A-containing mRNAs were analyzed using ribosome profiling datasets from HeLa cells as described in (A). Shown are the mean fold changes in TE (siMETTL3/siControl) for mRNAs with m6A residues only in the 5′ UTR (red), within the 3′ UTR (purple), within 50 nt of the stop codon (yellow), within the CDS and/or 3′ UTR (green), or in all mRNAs (blue), as defined by single-nucleotide-resolution m6A mapping. mRNAs with 5′ UTR m6A residues exhibit a dramatic reduction in TE after METTL3 depletion, whereas transcripts with m6As in other regions fail to show this effect. All mean fold change TE values were computed after background subtraction of the mean fold change computed from all nonmethylated control mRNAs, as indicated by the arrow (mean ± SEM; *p < 0.05).
(C) Fto knockdown increases heat-shock-induced translation of Hsp70. MEF cells stably expressing either Fto shRNA or scramble shRNA were subjected to heat shock stress. Cell lysates were collected at various times post-heat shock (“Post HS”) and then used for western blot analysis with the indicated antibodies. Fto knockdown increased the levels of stress-induced Hsp70 protein compared to control shRNA (“S exp” = short exposure; “L exp” = long exposure). Levels of Hsp25, another heat shock-induced protein, were unaffected by Fto knockdown. Right panel shows quantification of Hsp70 levels normalized to β-actin (n = 3; mean ± SEM; **p < 0.1).
(D) MEFs stably expressing control or Fto shRNA were subjected to heat shock stress as in (C). Polysome fractions were separated using sucrose gradient fractionation (left panels) followed by RT-qPCR for Hsp70 (top right panel) and Gapdh (bottom right panel) in each fraction. Hsp70 levels are increased in polysome fractions following Fto knockdown, whereas the distribution of Gapdh is unchanged (n = 3; mean ± SEM; Hsp70: p = 0.0007, two-way ANOVA; Gapdh: p = 0.3722, two-way ANOVA considering the entire range of time points).
(E) MEF cells were infected with either GFP or Fto lentivirus and subjected to heat shock stress. Cell lysates were collected at various times post-heat shock and then used for western blot analysis with the indicated antibodies. Fto overexpression decreased the levels of heat-shock-induced Hsp70 protein compared to GFP overexpression. Levels of Hsp25 were unaffected by Fto overexpression. Right panel shows quantification of Hsp70 levels normalized to β-actin (n = 3; Mean ± SEM; *p < 0.5).
(F) MEFs with or without Fto overexpression were subjected to heat shock stress as in (E). Polysome fractions were separated using sucrose gradient fractionation (left panels) followed by RT-qPCR of Hsp70 (top right panel) and Gapdh (bottom right panel) in each fraction. Hsp70 levels are decreased in polysome fractions following Fto overexpression, whereas the distribution of Gapdh is unchanged (n = 3; mean ± SEM; Hsp70: p < 0.0001, two-way ANOVA; Gapdh: p = 0.1910, two-way ANOVA considering the entire range of time points).
See also Figures S5 and S6 and Table S1.
Heat-Shock-Induced Translation of Hsp70 Is Mediated by 5′ UTR m6A
We next sought to investigate the role of m6A in promoting cap-independent translation in cells. Since cellular translation involves both cap-dependent and cap-independent mechanisms, we took advantage of heat shock, which induces a stress response that suppresses most cap-dependent translation (Holcik and Sonenberg, 2005). Heat-shock protein 70 (HSP70) is a stress response mRNA known to undergo increased transcription and cap-independent translation following heat shock (Lindquist and Craig, 1988). Previous studies demonstrated that HSP70 contains an m6A site within its 5′ UTR (Schwartz et al., 2014) and that methylation of the HSP70 5′ UTR is increased following heat shock (Dominissini et al., 2012). However, the role of m6A in cap-independent translation of HSP70 is not understood.
To test the effect of m6A in HSP70 translation, we utilized altered expression of Fto to influence m6A levels within the Hsp70 5′ UTR. Knockdown of Fto resulted in increased m6A levels in Hsp70 mRNA in heat-shocked cells (Figure S6A). Conversely, overexpressing Fto in heat-shocked cells reduced the level of m6A in Hsp70 mRNA by 29% relative to heat-shocked cells overexpressing GFP (Figure S6A). To determine whether altered m6A levels in the Hsp70 5′ UTR influence heat shock-induced Hsp70 translation, we used mouse embryonic fibroblasts (MEFs), which exhibit low Hsp70 levels prior to heat shock (Sun et al., 2011). In MEF cells stably expressing control shRNA, Hsp70 protein was readily detected 4 and 6 hr after heat shock. However, in MEF cells stably expressing _Fto_-specific shRNA to increase m6A levels, Hsp70 protein expression was significantly higher at both 4 and 6 hr after heat shock (Figure 6C). This effect was not due to increased levels of Hsp70 mRNA (Figure S6B). Furthermore, knockdown of Fto caused a significant increase in the fraction of polysome-bound Hsp70 mRNA (Figure 6D), suggesting that the increased levels of Hsp70 protein seen after heat shock reflect increased translation of Hsp70 mRNA in Fto knockdown cells.
Consistent with the effects of Fto knockdown on Hsp70 levels, Fto overexpression caused significantly reduced Hsp70 protein production 4 and 6 hr after heat shock (Figure 6E). This effect was not due to reduced Hsp70 transcript levels (Figure S6B). In addition, Hsp70 mRNA was significantly reduced in the polysome fractions of Fto-overexpressing cells compared to GFP-expressing cells, confirming that the Fto-mediated reduction in Hsp70 protein levels was due to reduced Hsp70 translation (Figure 6F). These data suggest that the loss of m6A in Hsp70 mRNA results in reduced translation efficiency following heat shock.
Transcriptome-wide Redistribution of m6A following Cellular Stress
We next sought to further understand the importance of 5′ UTR m6A residues in response to cellular stress. Based on our findings with Hsp70 mRNA, we considered the possibility that heat shock may alter the transcriptome-wide distribution of m6A. Under basal conditions, most m6A residues are located in mRNAs near the stop codon, with markedly fewer m6A residues in 5′ UTRs. To determine if cellular stress alters the characteristic distribution of m6A, we mapped m6A residues using miCLIP, a method for single-nucleotide resolution detection of m6A sites (Linder et al., 2015). Remarkably, the metagene analysis showed a marked enrichment of m6A in the 5′ UTR in heat-shocked cells compared to control cells (Figure S6C).
To further examine this phenomenon, we analyzed existing transcriptome-wide m6A mapping datasets that were performed in stressed cells and control cells. These include HepG2 cells treated with UV, interferon-γ, and heat shock (Dominissini et al., 2012). Metagene analyses showed prominent increases in the level of 5′ UTR m6A in both the UV-treated and heat-shocked cells (Figure S6D). The number of m6A sites in the 3′ UTR was relatively unaffected following heat shock or UV compared to control (n = 4,538, 4,533, or 3,171, respectively), whereas the number of m6A sites in the 5′ UTR was markedly increased in heat shock and UV relative to control (n = 1,501, 1,212, or 326, respectively) (Table S1). Notably, interferon-γ treatment did not alter the m6A metagene profile (Figure S6D), indicating that the induction of 5′ UTR m6A is not a nonspecific stress response but instead is linked to specific forms of cellular stress.
Intriguingly, both heat shock and UV caused increased 5′ UTR methylation in mRNAs that belong to common functional pathways, including phosphorylation and cell-cycle regulation (Table S1). Collectively, our results indicate that activation of some stress-response pathways causes a global reshaping of the cellular mRNA methylome and suggest that increased 5′ UTR methylation may be a general component of the response to select cellular stresses. Future studies will be important for understanding how stress pathways increase m6A within the 5′ UTR of mRNAs and reshape the RNA methylome. Furthermore, it will be important to analyze how diverse stress response pathways utilize these upregulated 5′ UTR m6A residues to mediate translational responses.
DISCUSSION
Eukaryotic mRNAs can be translated in both cap-dependent and cap-independent modes, although the mechanisms of translation initiation that do not require the 5′ cap and eIF4E have been poorly understood. Our results show that m6A residues within the 5′ UTR can act as an m6A-induced ribosome engagement site (MIRES), which promotes cap-independent translation of mRNA. We find that a single m6A in the 5′ UTR of mRNAs is sufficient to promote MIRES activity in cell-free extracts, whereas m6A residues outside the 5′ UTR fail to show this effect. The significance of 5′ UTR m6A residues is further seen in both ribosome profiling datasets and in individual cellular mRNAs in conditions where cap-dependent translation is suppressed. These results point to selective recognition of 5′ UTR m6A as a mechanism for mRNAs to bypass the cap requirement for translation and suggest a potential role for this class of m6A residues in mediating translational responses induced in diverse cellular stress states.
A role for m6A in promoting translation initiation is supported by our finding that METTL3 depletion leads to a large reduction in translation efficiency of mRNAs containing 5′ UTR m6A residues compared to mRNAs that contain m6As elsewhere. Although cap-independent translation of cellular mRNAs may also be mediated by m6A-independent pathways, including direct recruitment of ribosomes to internal 5′ sequence or structural elements (Xue et al., 2015), our studies raise the intriguing possibility that an eIF4E-independent mode of translation initiation can be switched on or off by reversible methylation of adenosine residues in the 5′ UTR of mRNAs.
Our studies show that cap-independent translation mediated by m6A requires a novel m6A reader, eIF3. We find that many eIF3-binding sites in the transcriptome overlap with m6A sites in 5′ UTRs. The identification of eIF3 as an m6A reader was originally suggested by the finding that the 48S complex can be assembled on m6A-containing RNA using only eIF1, eIF1A, eIF2, eIF3, and the 40S subunit. Of these components, eIF3 shows selective interaction with m6A both in vitro and in cells. By binding eIF3, 5′ UTR m6A residues can stimulate translation initiation by directly recruiting the 43S preinitiation complex to the 5′ UTR of mRNAs.
m6A has diverse effects on mRNAs, including mRNA destabilization and translational enhancement, although these effects are predominantly attributed to m6A near stop codons or in 3′ UTRs (Wang et al., 2014a, 2015). In the case of m6A near stop codons or in 3′ UTRs, translational enhancement is mediated by YTHDF1, which binds to select transcripts at m6A sites in their 3′ UTRs (Wang et al., 2015). YTHDF1 binds numerous proteins, including eIF3 and other ribosome-associated proteins, which are proposed to be recruited to 3′ UTRs to influence cap-dependent translation initiation (Wang et al., 2015). This is in contrast to the mechanism of 5′ UTR m6A, which directly recruits eIF3 and assembles translation initiation complexes in the 5′ UTR without cap-binding proteins. Our analysis of ribosome profiling data from YTHDF1-depleted cells further indicates that 5′ UTR m6A residues promote translation through a YTHDF1-independent mechanism. Thus, m6A exhibits markedly distinct effects on mRNA based on its location in transcripts.
A long-standing question is the mechanism by which select cellular mRNAs undergo cap-independent translation during conditions where cap-dependent translation is suppressed (Holcik and Sonenberg, 2005). A prevailing hypothesis has been that these mRNAs contain cellular IRESs that promote cap-independent translation (Komar and Hatzoglou, 2011). However, putative cellular IRESs often lack the complex structural elements seen in viral IRESs (Hellen and Sarnow, 2001). As a result of this discrepancy, and because of flaws inherent to many assays that test cellular IRES function, the evidence for and against cellular IRESs is a frequent topic of debate (Gilbert, 2010; Kozak, 2005). Given the prevalence of m6A within 5′ UTRs, their translation-promoting activity represents an additional or perhaps alternative mechanism for mediating cap-independent translation.
The importance of 5′ UTR m6A residues is supported by their selective upregulation in response to specific forms of stress. This m6A stress response points to the importance of this subset of m6A residues, which our results show are linked to cap-independent translation. Notably, other forms of stress regulate translation through the integrated stress response (Ron, 2002). It will be important to determine if 5′ UTR m6A-mediated translation is an alternative mechanism to orchestrate translational responses to stress.
EXPERIMENTAL PROCEDURES
In Vitro Translation
In vitro translation assays were performed using HeLa cell extracts (One-Step Human IVT Kit, Thermo Scientific). Equal amounts of RNA were used for each reaction (100 ng RNA per reaction, ~30 nM per reaction), and all reactions within each experiment were performed in equal volumes. Multiple different batches of HeLa extracts and mRNA preparations were used to ensure that the translation-promoting effect of m6A is not due to a specific lot of extract or batch of synthesized mRNA. However, this also contributes to inter-experiment variability. Reactions were performed at 30°C for 30 min and were stopped by the addition of 200 μM cycloheximide and placed on ice. 1 μl of each reaction was then used for luminescence analysis (see below). The remaining reaction volume was used for RNA isolation with TRIzol (Invitrogen) or QIAGEN RNeasy kits according to the manufacturer’s instructions. cDNA synthesis was then performed using Superscript III reverse transcriptase (Invitrogen) and random hexamers. Following treatment with RNase H, cDNA was then used for RT-qPCR analysis to ensure that differences in mRNA levels across samples did not account for the observed changes in protein production. Statistical analysis of luciferase activity measurements (below) was performed using Student’s t test and a p value threshold of 0.01.
Luciferase Activity Measurements
Luciferase expression was measured using the One-Glo luciferase assay kit (Promega) according to the manufacturer’s instructions. Luminescence measurements were performed on a Molecular Devices Spectramax L micro-plate reader using the SoftMax Pro software program.
eIF3a PAR-iCLIP
eIF3a PAR-iCLIP was performed using HEK293T cells as described previously (Huppertz et al., 2014) with some adjustments. 10 million cells were incubated with 100 mM 4SU for 8 hr. Media was then discarded, and cells were placed on ice and irradiated with 365 nm UV light using a Stratalinker UV crosslinker (Stratagene) with 150 mJ/cm2. Cells were scraped in ice-cold 1×PBS and collected by centrifugation at 200 x g for 10 min at 4°C. Cell pellets were suspended in 200 μl of 1% SDS, 10 mM DTT, and 1×protease inhibitors (cOmplete mini EDTA-free, Roche). The lysate was then passed through an 18G needle 10 times to improve cell lysis and shearing of DNA. SDS was neutralized by diluting the lysate to 2 ml using RIPA buffer without SDS. The remainder of the protocol was performed as described (Huppertz et al., 2014) using rabbit anti-eIF3a (Abcam).
Additional methods are detailed in the Supplemental Experimental Procedures section.
Supplementary Material
1
2
3
4
5
6
7
8
9
Highlights.
- m6A residues within the 5′ UTR promote cap-independent translation
- Translation of cellular mRNAs is increased by the presence of m6A within the 5′ UTR
- Heat shock induces Hsp70 translation in an m6A-dependent manner
- Diverse cellular stresses increase 5′ UTR adenosine methylation
Acknowledgments
We thank members of the S.R.J. laboratory for helpful comments and suggestions. This work was supported by NIH grant K99MH104712 and the Revson Senior Fellowship in Biomedical Sciences (K.D.M.), NIH grants R01DA037755 (S.R.J.), AG042400 (S.-B.Q), and GM59660 (T.V.P.), and NSF CAREER grant 1054964 (O.E.).
Footnotes
ACCESSION NUMBERS
Sequencing data have been deposited to NCBI’s GEO under accession number GEO: GSE73405.
AUTHOR CONTRIBUTIONS
K.D.M., T.V.P., S.-B.Q., and S.R.J. designed the experiments and analyzed data. J.Z. and S.-B.Q. performed experiments related to Fto and Hsp70. K.D.M. and S.R.J. wrote the manuscript. K.D.M., D.P.P., J.Z., A.Z., and M.A.S. performed the experiments, collected data, and prepared the figures. O.E. and D.P.P. performed computational analysis of sequencing data. All authors commented and made edits to the manuscript.
References
- De Gregorio E, Preiss T, Hentze MW. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–495. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010;285:29033–29038. doi: 10.1074/jbc.R110.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse S, Zhong S, Bodi Z, Button J, Alcocer MJ, Hayes CJ, Fray R. A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci Rep. 2011;1:126. doi: 10.1038/srep00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Cate JH. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami S, Masutani M, Sonenberg N, Yokoyama S, Imataka H. An efficient mammalian cell-free translation system supplemented with translation factors. Protein Expr Purif. 2006;46:348–357. doi: 10.1016/j.pep.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PS, Grover R, Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006;7:404–410. doi: 10.1038/sj.embor.7400623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D. Translational control in the endoplasmic reticulum stress response. J Clin Invest. 2002;110:1383–1388. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Correction to “Structure and Thermodynamics of N(6)-Methyladenosine in RNA: A Spring-Loaded Base Modification”. J Am Chem Soc. 2015;137:8308. doi: 10.1021/jacs.5b05858. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Sun J, Conn CS, Han Y, Yeung V, Qian SB. PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J Biol Chem. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Andreev DE, Dmitriev SE, Shatsky IN. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 2013;41:1807–1816. doi: 10.1093/nar/gks1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014b;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7
8
9