RNA sensors of the innate immune system and their detection of pathogens (original) (raw)
Summary
The innate immune system plays a critical role in pathogen recognition and initiation of protective immune response through the recognition of pathogen associated molecular patterns (PAMPs) by its pattern recognition receptors (PRRs). Nucleic acids including RNA and DNA have been recognized as very important PAMPs of pathogens especially for viruses. RNA are the major PAMPs of RNA viruses, to which most severe disease causing viruses belong thus posing a tougher challenge to human and animal health. Therefore, the understanding of the immune biology of RNA PRRs is critical for control of pathogen infections especially for RNA virus infections. RNA PRRs are comprised of TLR3, TLR7, TLR8, RIG‐I, MDA5, NLRP3, NOD2, and some other minorities. This review introduces these RNA PRRs by describing the cellular localizations, ligand recognitions, activation mechanisms, cell signaling pathways, and recognition of pathogens; the cross‐talks between various RNA PRRs are also reviewed. The deep insights of these RNA PRRs can be utilized to improve anti‐viral immune response. © 2017 IUBMB Life, 69(5):297–304, 2017
Keywords: pattern recognition receptors (PRRs), RNA, pathogen associated molecular patterns (PAMPs), pathogens
Abbreviations
PAMPs
pathogen associated molecular patterns
PRRs
pattern recognition receptors
TLRs
Toll‐like receptors
RIG‐I
retinoic acid inducible gene‐I
MDA5
melanoma differentiation‐associated gene 5
NOD2
nucleotide‐binding and oligomerization domain containing 2
NLRP3
NOD‐like receptor, pyrin domain containing 3
AIM2
absent in melanoma 2
IFI16
interferon‐inducible protein 16
RLRs
RIG‐I like receptors
NLRs
NOD‐like receptors
CLRs
C‐type lectin receptors
ALRs
AIM2‐like receptors
cGAS
cyclic GMP‐AMP synthase
DAMPs
damage associated molecular patterns
MHC
major histocompatibility complex
RNA
ribonucleic acid
DNA
deoxyribonucleic acid
ECD
ectodomain or extracellular domain
TM
transmembrane domain
TIR
cytoplasmic Toll/IL‐1 receptor
LRR
leucine rich repeats
TRIF
TIR domain‐containing adaptor‐inducing interferon‐β
MyD88
myeloid differentiation primary response 88
NF‐κB
nuclear factor kappa B
IRF
interferon regulatory factor
pDCs
plasmacytoid dendritic cells
dsRNA
double‐stranded RNA
TRAF3
TNF receptor associated factor 3
TBK1
TANK binding kinase 1
IKK
IκB‐Kinase
ssRNA
single‐stranded RNA
siRNA
small interfering RNA
miRNA
micro RNA
IRAK
interleukin‐1 receptor‐associated kinase
IFN
interferon
CARDs
caspase recruitment domains
RD
repressor domain or regulatory domain
MAVS
mitochondrial antiviral signaling protein
NEK7
NIMA related kinase 7
ROS
reactive oxygen species
ASC
apoptosis‐associated speck‐like protein containing a CARD domain
IL
interleukin
RIP2
receptor‐interacting protein 2
IAV
influenza A virus
DHX
DExH‐box helicase
DDX
DExD/H‐box helicase
LRRFIP1
leucine‐rich repeat flightless‐interacting protein 1
HIV
human immunodeficiency virus
FMDV
foot‐mouth disease virus
CMV
cytomegalovirus
HSV‐1
herpes simplex virus‐1
EMCV
encephalomyocarditis virus
WNV
west nile virus
SeV
sendai virus
MV
measle virus
RSV
respiratory syncytial virus
EBOV
ebola virus
NDV
newcastle disease virus
VSV
vesicular stomatitis virus
RV
rabies virus
HCV
hepatitis C virus
JEV
Japanese encephalitis virus
SGB
streptococcus group B.
Introduction
The innate immune system represents the first line of defense against pathogens through its continuous monitoring of the pathogen associated molecular patterns (PAMPs), and subsequent activation of a series of defense mechanisms to eliminate the infections. The concept and model of innate immune sensing was first proposed by Charles Janeway Jr. who predicted that there must exist a group of innate immune receptors responsible for recognition and sensing of non‐self from self, and triggering subsequent adaptive immunity 1. Later studies confirmed his prediction and more and more innate immune receptors called pattern recognition receptors (PRRs) have been found since then.
Based on protein domain homology, PRRs have been divided into several families; they are Toll‐like receptors (TLRs), RIG‐I like receptor (RLRs), NOD‐like receptors (NLRs), C‐type lectin receptors (CLRs), AIM2‐like receptors (ALRs), and the recently discovered cytosol DNA sensing PRR cyclic GMP‐AMP synthase (cGAS). These PRRs recognize and sense a variety of PAMPs from viruses, bacteria, fungi, and protozoa, which range from lipoproteins, carbohydrates, lipopolysaccharide to nucleic acids. PRRs also recognize endogenous damage associated molecular patterns (DAMPs) from host, which is related with both homeostasis and autoimmune diseases. Upon sensing of PAMPs or DAMPs, the PRRs trigger intracellular cell signaling, leading to transcriptional activation and expression of cytokines, chemokines, MHC, and co‐stimulatory molecules. Additionally, PRR triggered cell signaling induces several transcription‐independent cell processes such as phagocytosis, autophagy, cell death, and inflammasome/cytokine processing, which work together with the transcriptional innate responses 2.
The nucleic acids RNA and DNA have drawn much attention as important PAMPs 3, 4. Different from non‐pathogens, the pathogens including viruses and intracellular bacteria replicate in cells, and nucleic acids RNA or DNA represent the signature of pathogens in particular of viruses which accumulate large amount of nucleic acids during replication in cells. All the nucleic acid detecting PRRs are localized intracellularly. For example, DNA sensing PRRs are endosomal TLR9, cytosolic AIM2, IFI16, and cGAS; RNA sensing PRRs are endosomal TLR3, TLR7, TLR8, and cytosolic RIG‐I, MDA5, NLRP3, and NOD2. RNA PRRs play more important roles than DNA PRRs in recognition of RNA virus infections and initiation of protective immune responses. RNA viruses exhibit rapid replication kinetics, high mutation rates, and complex evolutionary dynamics, thus RNA viruses pose unique challenges to human and animal health 5. Most severe disease causing viruses are RNA viruses, such as ebola virus, influenza virus, human immunodeficiency virus (HIV), foot‐mouth disease virus (FMDV), etc. Therefore, investigation and understanding of RNA PRRs are critical for control of virus infections and protection of host. Following are the description of individual RNA PRR.
TLRs: TLR3, TLR7, and TLR8
The family of Toll like receptors (TLRs) are the earliest discovered PRRs. Currently human and mouse have 10 and 12 TLRs, respectively: both human and mouse have TLR1‐9; in addition, human has TLR10 whereas mouse has TLR11‐13 6. All TLRs are type I transmembrane proteins and comprised of N‐terminal ectodomain or extracellular domain (ECD), middle transmembrane domain (TM) and C‐terminal cytoplasmic Toll/IL‐1 receptor (TIR) domain (Supporting Information Fig. 1). The ECD contains 20–26 leucine rich repeats (LRR) motifs, which are juxtaposed into a horseshoe‐shaped solenoid or a ring‐like structure. The α‐helix of each LRR is located on the convex surface of solenoid structure, and β‐sheet of each LRR assembles and forms into the concave surface of the solenoid structure. Different from other LRR containing proteins, TLRs bind their ligands including agonists on the lateral convex surface instead of concave surface 7. The formation of M‐shaped dimer or multimer is needed for all TLR activation, so that the C‐terminal regions of the two TLR ECDs are brought into proximity. It in turn causes the multimerization of cytoplasmic TIR domains, which will recruit downstream adaptors TRIF or MyD88 through homotypic interaction, further forming signaling complex called signalosome and activating downstream transcription factors: one is NF‐κB that induces proinflammtory cytokines, another is interferon regulatory factor (IRF) that induces anti‐viral type I Interferon (IFN) 6.
TLR3 is widely distributed in all innate immune cells except neutrophils and plasmacytoid dendritic cells (pDCs), and localized in the endosomes of these cells 8. TLR3 recognizes the double‐stranded RNA (dsRNA) of viruses and the synthetic dsRNA analog poly I:C 9. The crystal structure of TLR3 ECD was the first resolved one among TLR proteins, existing as a monomer of solenoid structure. Upon binding to dsRNA, TLR3 forms a dimer with the backbone phosphates and sugars of dsRNA binding to the lateral positive charged regions of N‐ and C‐terminal ECD 10. The downstream adaptor TRIF is recruited by activated TLR3 through homotypic interaction, which further forms TRIF signaling complex involving other signaling components, such as TRAF6, TRAF3, TBK1, IKKε, and IKK. The transcription factors IRF3/IRF7 and NF‐κB are subsequently activated by the signaling complex and induce the expression of IFN and proinflammatory cytokines, respectively (Fig. 1).
Figure 1.
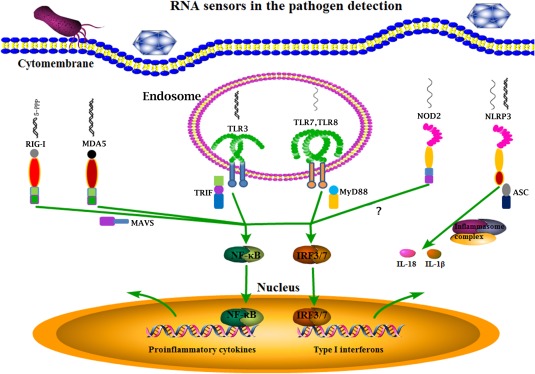
The RNA sensors in pathogen detection. The RNA viruses and some bacteria can be recognized by the main RNA sensors inside cells including endosomal TLR3, TLR7, TLR8, cytosolic RIG‐I, MDA5, NLRP3, and NOD2. Except NLRP3 which initiates inflammasome formation and processes cytokines for maturation, all others activate downstream NF‐κB and IRF signaling pathways, and thus cytokine gene transcription. All these RNA sensor induced signaling together combats pathogen infections.
TLR3 recognizes the genomic dsRNA of reoviruses, and the intermediate RNAs generated during replication of different viruses including mouse cytomegalovirus (MCMV), herpes simplex virus‐1 (HSV‐1), encephalomyocarditis virus (EMCV), flaviviruses, and enteroviruses (Table 1). In these cases, the activated TLR3 signaling restricts the virus replication; however, the TLR3 induced inflammation also contributes to the breaching of blood–brain barrier, leading to neuropathologies exemplified as West Nile Virus (WNV) infection in mice 11, 12.
Table 1.
The summary of cellular localizations and distributions, ligand recognitions, activation mechanisms, cell signaling, recognition of pathogens, and cross‐talks for RNA PRRs
| TLR3 | TLR7 | TLR8 | RIG‐I | MDA5 | NLRP3 | NOD2 | |
|---|---|---|---|---|---|---|---|
| Cellular localizations, distributions | Endosomes. Innate immune cells except neutrophils and pDCs | Endosomes. pDCs and B cells | Endosomes. Monocytes, macrophages and cDCs | Cytoplasm. All mammalian cell types | Cytoplasm. All mammalian cell types | Cytoplasm. Ubiquitously expressed | Cytoplasm. Macrophages, monocytes, Paneth cells, DCs |
| Recognized ligands | dsRNA, and the synthetic dsRNA analog poly I:C | R837, Loxoribine, R848, CL097, ssRNA | CL075, R848, CL097, ssRNA | 5' ppp‐dsRNA, short dsRNA | Long dsRNA | pathogen ssRNA/dsRNA and other distinct set of ligands | bacterial MDP and virus RNA |
| Activation mechanisms | dsRNA induced dimerization | Z‐loop proteolytic cleavage, and receptor dimer conformational change | Z‐loop proteolytic cleavage, and receptor dimer conformational change | K63‐polyubiquitination/polyubiquitin chain binding mediated receptor tetramerization | filament formation mediated receptor tetramerization | potassium efflux‐NEK7 involved NLRP3 inflammasome complex | Likely tetramerization into washer‐locker structure |
| Cell signaling pathways | TRIF‐TRAF3‐TBK1/IKKε‐IRF3. TRIF‐TRAF6‐IKKs‐NF‐κB | MyD88‐IRAK4/IRAK1‐IRF7. MyD88‐IRAF6‐IKKs‐NF‐κB | MyD88‐IRAK4/IRAK1‐IRAF6‐IKKs‐NF‐κB | MAVS‐TRAF3‐TBK1/IKKε‐IRF3. MAVS‐FADD/TRAF6‐IKKs‐NF‐κB | MAVS‐TRAF3‐TBK1/IKKε‐IRF3. MAVS‐FADD/TRAF6‐IKKs‐NF‐κB | ASC‐inflammasome‐caspase‐1‐IL‐1/IL‐18 | RIP2‐IKKs‐NF‐κB |
| Recognized pathogens | MCMV, HSV‐1, EMCV, WNV, and enteroviruses | SeV, flu virus, coxsackie virus, vaccinia virus, MV, RSV, retrovirus; SGB | SeV, flu virus, coxsackie virus, vaccinia virus, MV, RSV, retrovirus; E. coli, M. bovis, H. pylori, B. burgdorferi | EBOV, MV, SeV, NDV, RSV, flu virus, hantavirus, VSV, RV, HCV, JEV, adenovirus, vaccinia virus, HSV, L. monocytogenes, H. pylori, S. flexneri; Rotavirus, dengue virus, WNV, murine hepatitis virus | EMCV, poliovirus and coxasackie virus; Rotavirus, dengue virus, WNV, murine hepatitis virus | flu virus, SeV and bacteria | RSV, IAV, and HCMV |
| Cross‐talks | TLR8 (+) | NOD2 (+) | TLR3 (+); TLR7 (−); NOD2 (+) | TLR3 (−); NOD2 (−) | TLR3 (−) | RIG‐I (−) |
TLR7 and TLR8—belong to the TLR7/8/9 subfamily whose members are all localized at endosomes of the cells. TLR7 and TLR8 are very similar in terms of ligand recognition and intracellular signaling. Both are activated by small molecular agonists and GU or U rich single‐stranded RNA (ssRNA) (Fig. 1). Small molecular compounds Resiquimod (R848) and CL097 are agonists for both TLR7 and TLR8; however, Imiquimod (R837) and Loxoribine only for TLR7, and CL075 (3M002) only for TLR8. In addition, TLR7 and TLR8 may also recognize short dsRNA such as siRNA from RNA interference (RNAi), and some miRNA such as miRNA‐21 and miRNA‐29a secreted by tumor cells 13. The ECDs of TLR7 and TLR8 both have 26 LRR motifs that are more than those of TLR1‐6; additionally there are several insertions including the one between LRR14 and LRR15 called undefined region or Z‐loop, therefore, the ECDs are larger and exhibit the ring‐like crystal structures 14. The TLR8 and TLR7 at endosome are proteolytically cleaved along the Z‐loop by cathepsins and arginine endopeptidase; nevertheless, the two cleaved fragments are still stuck together by multiple intermolecular interactions, and both are required for receptor's activation 15, 16. The Z‐loop cleavage is necessary for TLR8/7 dimerization which is essential for activation 17. Newly recent crystal structures showed that there are dual agonist binding sites in both TLR7 and TLR8 dimers 18, 19: one is located within dimer interfaces which binds small chemical agonists or degrade products of ssRNA agonists; another is located on the concave surface of the TLR horseshoe structures and for binding of ssRNA oligonucleotides. The first sites are enough for small chemical agonist induced TLR7 and TLR8 activation, whereas both sites are necessary for ssRNA induced TLR7 and TLR8 activation. At steady state, TLR7 and TLR8 exist as dimers; upon binding to agonists, the conformation of dimers change such that the cytoplasmic TIR domains multimerize and recruit downstream adaptor MyD88 through homotypic interaction. The signaling complex called Myddosome is formed involving IRAK4, IRAK1, TRAF6, TRAF3 and downstream transcription factors NF‐κB and IRF7 are activated to induce proinflammatory cytokines and IFNs, respectively.
Despite the high similarity between TLR7 and TLR8, the cell distributions of these two are almost opposite: TLR7 is exclusively expressed by plasmacytoid dendritic cells (pDCs) and B cells, whereas TLR8 is expressed mainly in monocytes, macrophages and conventional dendritic cells (cDCs), very low level in pDCs and B cells 20. Both TLR7 and TLR8 are able to recognize multiple virus infections, including sendai virus (SeV), influenza virus, coxsackie virus, vaccinia virus, measle virus (MV), respiratory syncytial virus (RSV), and retrovirus 11. Furthermore, TLR7 recognizes Streptococcus Group B (SGB) RNA 21, 22, and TLR8 recognizes the RNAs from Escherichia coli, Mycobacteria bovis, Helicobacter pylori, and Borrelia burgdorferi 23 (Table 1).
RLRs: RIG‐I and MDA5
RLRs are expressed in almost all mammalian cell types, and as the main family of cytosolic RNA sensors play key roles in virus recognition and immune responses 24, 25. The prototypic RIG‐I was first discovered through screening of cDNA library 26, 27, other members of this family include MDA5 and LGP2. RIG‐I and MDA5 have similar domain structures: N‐terminal effective two tandem caspase recruitment domains (2CARDs), middle DExH‐box helicase domain and C‐terminal repressor domain, or regulatory domain (RD) 24 (Supporting Information Fig. 1). The optimal RNA recognized by RIG‐I is the 5′ ppp‐dsRNA, whereas the one MDA5 prefers is long dsRNA (Fig. 1). The third member LGP2, lacks of N‐terminal 2CARDs, has no signaling activity, but is able to regulate RIG‐I and MDA5 signaling due to the capability of binding RNA 24, 25.
RIG‐I, as the prototypic member of RLRs, has been subjected to extensive research. Currently, there have been intensive investigations and clear understanding of RIG‐I activation mechanism: Under steady state, RIG‐I CARDs binds to the Hel‐2i region of the helicase and is subjected to auto‐inhibition 25. Upon binding of RIG‐I C‐terminal RD to RNA, the conformation of RIG‐I changes so that the RNA further binds to Hel‐2i, and in turn the auto‐inhibition of 2CARDs is released. Next, through K63‐polyubiquitination/polyubiquitin chain binding of the 2CARDs and/or the filament formation by RIG‐I RD‐helicase along the dsRNA chain, the 2CARDs are tetramerized into stable lock‐washer structure. The effective 2CARD tetramer will nucleate downstream adaptor MASV, which aggregates on the mitochondria into prion‐like signaling complex 28. The downstream TRAF3/TBK1/IKKε and TRAF6/IKK further activate transcription factors IRF3 and NF‐κB, which drive IFN and proinflammtory cytokine expression, respectively 29 (Fig. 1).
Consistent with the recognition of short dsRNA ligands, RIG‐I specifically recognizes most single‐negative RNA viruses which generate lots of short 5′ ppp‐dsRNA during replication. These viruses include but are not limited to ebola virus (EBOV) of filovirus family, measle virus (MV), sendai virus (SeV), newcastle disease virus (NDV), respiratory syncytial virus (RSV) of paramyxovirus family, influenza virus of othomyxovirus family, hantavirus of bunyavirus family, vesicular stomatitis virus (VSV), and rabies virus (RV) of rhabdovirus family. RIG‐I also recognizes positive single RNA viruses such as hepatitis C virus (HCV) and Japanese encephalitis virus (JEV) of flavivirus family. In addition, RIG‐I is able to sense some DNA viruses such as adenovirus, vaccinia virus, herpes simplex virus (HSV) in that these viruses produce small dsRNA through their type III RNA polymerase during replication. There was also report showing that RIG‐I can detect the RNA from bacteria such as Listeria monocytogenes, Helicobacter pylori, and Shigella flexneri 30.
MDA5 has a very similar activation mechanism to that of RIG‐I: RD‐helicase binds with RNA, and conformational change exposes the N terminal 2CARDs, which form tetramer structure. Because MDA5 binds long dsRNA, the 2CARD tetramer is stabilized mainly through the filament formation along the dsRNA, and thus less dependent on K63‐polyubiquitination/polyubiquitin chain binding 28. The effective 2CARD tetramer will nucleate downstream adaptor MAVS, activate TRAF3/TBK1/IKKε/IRF3 and TRAF6/IKK/NF‐κB, which drive IFN and proinflammtory cytokine expression, respectively 29.
MDA5 recognizes long dsRNA, and accordingly senses the single positive RNA viruses such as the encephalomyocarditis virus (EMCV), poliovirus and coxasackie virus of picornavirus family 30. On the other hand, both RIG‐I and MDA5 cross‐detect the same viruses; these viruses are double RNA rotavirus of reovirus family, dengue virus, and west nile virus (WNV) of flavivirus family, murine hepatitis virus of coronavirus family 30 (Table 1).
NLRs: NLRP3 and NOD2
NLR is also the cytosolic receptor family, which is mainly utilized for detection of bacteria. Human has 22 members, whereas mouse has 34 members. All NLR members have similar domain structures: N‐terminal effector domain, middle nucleotide‐binding and oligomerization domain (NOD) and C‐terminal leucine rich repeats (LRRs) (Supporting Information Fig. 1). Based on the N‐terminal effector domains, the NLR is divided into five subfamilies: NLRA (Acid activation domain), NLRB (Baculovirus inhibitor of apoptosis repeats), NLRC (Caspase activation and recruitment domain CARD), NLRP (Pyrin domain PYD), and NLRX (Unknown domain) 31. Except NOD1 and NOD2, all the NLRs upon activation induce the inflammasome formation other than gene transcription.
NLRP3, as one of the most extensively studied NLRs, is able to recognize a very broad and distinct set of ligands including ATP, uric acid, silica, adjuvant aluminum, cholesterol crystals, nigericin, pore‐forming proteins, mitochondrial DNA, and pathogen mRNA. Common among these stimulators, NLRP3 is activated directly by potassium efflux coupled with downstream NEK7 32, 33. Nevertheless, there existed potassium efflux independent non‐canonical NLRP3 inflammasome formations under conditions of disrupted glycolytic flux or by stimulation of extracellular LPS 33. Probably, NLRP3 would favor a general sensor of homeostatic disruption such as lysosomal rupture, mitochondria damage, reactive oxygen species (ROS), and ionic imbalance 31. However, the exact mechanisms of NLRP3 action need to be further investigated and determined. Under steady state, NLRP3 is likely subjected to auto‐inhibition; upon activation, NLRP3 binds downstream adaptor ASC and substrate procaspase‐1, forming the wheel‐like structure inflammasome. Next the caspase‐1 is activated, processing the pro‐IL‐1 and pro‐IL‐18 into active IL‐1 and IL‐18 (Fig. 1). NLRP3 recognizes the cytosolic dsRNA/ssRNA of influenza virus and sendai virus, and bacteria mRNA, activating the inflammasome and inducing IL‐1 and IL‐18 34, 35, 36 (Table 1).
NOD2 and NOD1 (also called NLRC2 and NLRC1), different from other NLRs, recognize bacterial glycopeptides MDP and ie‐DAP, respectively, recruit downstream adaptor RIP2 through homotypic interaction forming signaling complex, and induce NF‐κB activation and proinflammatory cytokine expression 31. NOD2 receptors are found mostly in macrophages, monocytes, paneth intestinal cells, and dendritic cells 37. Additional studies suggested that NOD2 plays an important role in the restriction of respiratory syncytial virus (RSV), influenza A virus (IAV), and human cytomegalovirus (HCMV), likely through the recognition of virus RNA and subsequent IFN induction 38, 39 (Table 1). Similar to RLRs, NOD2 has N‐terminal 2CARDs, and may tetramerize into washer‐locker structure to initiate the adaptor MAVS aggregation and downstream IFN production 38, 40 (Fig. 1). To this end, the crystal structure of NOD2 CARDs has to be solved in the future 31.
Other Minor RNA PRRs
TLR13 is only expressed in mouse, and localized in the endosome compartment. Once activated by RNA, TLR13 recruits adaptor MyD88 to trigger transcription factor NF‐κB activation and induce proinflammtory cytokine production 22. Mouse TLR13 was reported to recognize a conserved CGGAAAGACC motif in Staphylococcus aureus 23S rRNA and E. coli 23S rRNA 41, 42. Furthermore, TLR13 also recognizes Streptococcus pyogenes 43. In addition to RLRs RIG‐I and MDA5, there exists a group of non‐RLR RNA helicases which mediate cytosolic RNA recognition and signaling. These are DDX3, DHX9, DHX33, DDX60, and DDX1/DDX21/DHX36 30, 44. DDX3 binds poly I:C or vesicular stomatitis virus (VSV) RNA, associates with RLR‐MAVS signaling complex, and enhances IFN response. DHX9 is expressed in mouse spleen dendritic cells and mouse bone marrow dendritic cells, recognizes poly I:C, influenza virus and reovirus RNA, binds with MAVS activating downstream signaling. DHX33 binds poly I:C and reovirus RNA, activates MAVS mediated signaling or NLRP3 mediated inflammasome. DDX60 can be induced by the virus infection, and in the meantime binds the virus RNA, further binds RIG‐I, MDA5, and LGP2, enhancing RLR signaling and downstream IFN response. DDX1/DDX21/DHX36 are expressed in myeloid dendritic cells, in which DDX1 binds poly I:C through helicase domain, whereas DHX36 and DDX21 bind TRIF TIR domain through their HA2‐DUF and PRK domain, respectively. The complex enhances IFN response and exhibits inhibitory effect to influenza virus and reovirus infections. The third minor RNA PRR, leucine‐rich repeat flightless‐interacting protein 1 (LRRFIP1) was reported to bind both RNA and DNA and be involved in the recognition of vesicular stomatitis virus (VSV) and Listeria monocytogene, activate β‐catenin, which then translocates into nucleus and promotes IRF3 transcription activity and IFN production 45.
The Interactions Between RNA PRRs
Interactions of microbes with the innate immune system involve the parallel recognition of different PAMPs of the whole pathogen by multiple PRRs and simultaneous induction of multiple PRR signaling pathways, for which evidence has been accumulated in the past years 46, 47, 48. Likewise, there are interactions between different RNA PRRs and mutual influence of different RNA PRR signaling. Understanding of the enormous complexity of these processes helps provide insights into in vivo innate immune activation. Within the RNA sensing TLRs, synergistic effects were observed between TLR3 and TLR8 in monocyte‐derived macrophages and DCs 49, 50; In contrast, TLR8 inhibits TLR7 signaling in both human and mice 51, 52. Between three subfamilies of RNA PRRs, TLR7/8 cooperates with NOD2 in DC activation and results in a synergistic release of pro‐inflammatory mediators which promote the activation of Th17 cells 53. RLRs suppress the gene transcription of IL‐12p40 induced by the activation of Toll‐like receptors (TLRs) including TLR3 and subsequent TLR3 induced Th1 and Th17 responses 47. Furthermore, RLR RIG‐I cross‐interferes with NOD2 in regulating downstream signaling by direct interaction with each other 54. The interactions between different PRRs need to be considered in the contexts of distinct species, various cell types, and different agonists used for stimulation.
The Ability to Devise Strategies to Control Virus Infections will be Improved with the Knowledge of RNA PRRs
The understanding of RNA PRR immune biology including the ligand recognitions, cellular localizations, cell signaling pathways, mechanisms of activation, recognized pathogens and the interactions between different RNA PRRs will definitely be helpful to improve the anti‐viral immune response. We give some examples here: First, based on the ligand recognitions and activation mechanisms of RNA PRRs, RNA varieties and small‐molecular agonists of higher potency can be developed for either direct anti‐viral therapy or effective viral vaccine adjuvants. Similarly, elucidation of the RNA PRR triggered cell signaling pathways will potentially lead to the discovery of novel innate immune modulators with higher anti‐viral efficacy. Second, for certain virus infections, only the RNA PRRs involved in the viral recognition will likely play an important role in the anti‐viral immune response; in this case, knowledge of the virus recognition by RNA PRRs becomes critical. Third, TLR3, 7, 8 are primarily expressed by macrophages and DCs and recognize viral RNA within the endosomal compartment, whereas RLRs (RIG‐I, MDA5) and NLRs (NLRP3, NOD2) are ubiquitously expressed and sense viral RNA within the cytoplasm of infected cells (Table 1). Therefore, it should be considered how to deliver the RNA PRR agonists to host to maximize the anti‐viral immune response, such as delivery routes, with or without transfection etc. Fourth, as seen in Table 1, TLRs have general positive cross‐talks with NLRs, whereas RLRs have negative cross‐talks with TLRs and NLRs. Even though information is incomplete, this knowledge of cross‐talks between different RNA PRRs will provide a rationale to develop appropriate combinations of various agonists/modulators to control viruses which are simultaneously recognized in vivo by several RNA PRRs. With the information becoming complete, and additional knowledge on how the RNA PRR signaling influences downstream adaptive immune response, more and more effective therapeutics or vaccine adjuvants to control virus infections will be on the horizon.
Supporting information
Supporting Information Figure 1
Acknowledgments
The work was partly supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions and Jiangsu Co‐innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, China, by Natural Science Foundation of China (31672523) to J.Z. and by China Postdoctoral Science Foundation (2016M590510) to N.C.
References
- 1.Medzhitov, R. (2009) Approaching the asymptote: 20 years later. Immunity 30, 766–775. [DOI] [PubMed] [Google Scholar]
- 2.Brubaker, S. W. , Bonham, K. S. , Zanoni, I. , and Kagan, J. C. (2015) Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbalat, R. , Ewald, S. E. , Mouchess, M. L. , and Barton, G. M. (2011) Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 29, 185–214. [DOI] [PubMed] [Google Scholar]
- 4.Luecke, S. , and Paludan, S. R. (2016) Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine. In press. DOI: 10.1016/j.cyto.2016.10.003 [DOI] [PubMed] [Google Scholar]
- 5.Zhu, J. , Ghosh, A. , and Sarkar, S. N. (2015) OASL‐a new player in controlling antiviral innate immunity. Curr. Opin. Virol. 12, 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai, T. , and Akira, S. (2010) The role of pattern‐recognition receptors in innate immunity: update on Toll‐like receptors. Nat. Immunol. 11, 373–384. [DOI] [PubMed] [Google Scholar]
- 7.Werling, D. , Jann, O. C. , Offord, V. , Glass, E. J. , and Coffey, T. J. (2009) Variation matters: TLR structure and species‐specific pathogen recognition. Trends Immunol. 30, 124–130. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi, F. , Means, T. K. , and Luster, A. D. (2003) Toll‐like receptors stimulate human neutrophil function. Blood 102, 2660–2669. [DOI] [PubMed] [Google Scholar]
- 9.Edelmann, K. H. , Richardson‐Burns, S. , Alexopoulou, L. , Tyler, K. L. , Flavell, R. A. , et al. (2004) Does Toll‐like receptor 3 play a biological role in virus infections?. Virology 322, 231–238. [DOI] [PubMed] [Google Scholar]
- 10.Choe, J. , Kelker, M. S. , and Wilson, I. A. (2005) Crystal structure of human toll‐like receptor 3 (TLR3) ectodomain. Science 309, 581–585. [DOI] [PubMed] [Google Scholar]
- 11.Thompson, M. R. , Kaminski, J. J. , Kurt‐Jones, E. A. , and Fitzgerald, K. A. (2011) Pattern recognition receptors and the innate immune response to viral infection. Viruses 3, 920–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma, R. , and Bharti, K. (2017) Toll like receptor 3 and viral infections of nervous system. J. Neurol. Sci. 372, 40–48. [DOI] [PubMed] [Google Scholar]
- 13.Sarvestani, S. T. , Williams, B. R. , and Gantier, M. P. (2012) Human Toll‐like receptor 8 can be cool too: implications for foreign RNA sensing. J. Interferon Cytokine Res. 32, 350–361. [DOI] [PubMed] [Google Scholar]
- 14.Tanji, H. , Ohto, U. , Shibata, T. , Miyake, K. , and Shimizu, T. (2013) Structural reorganization of the Toll‐like receptor 8 dimer induced by agonistic ligands. Science 339, 1426–1429. [DOI] [PubMed] [Google Scholar]
- 15.Bauer, S. (2013) Toll‐like receptor 9 processing: the key event in Toll‐like receptor 9 activation?. Immunol. Lett. 149, 85–87. [DOI] [PubMed] [Google Scholar]
- 16.Ohto, U. , Tanji, H. , and Shimizu, T. (2014) Structure and function of toll‐like receptor 8. Microbes Infect./Instit. Pasteur 16, 273–282. [DOI] [PubMed] [Google Scholar]
- 17.Tanji, H. , Ohto, U. , Motoi, Y. , Shibata, T. , Miyake, K. , et al. (2016) Autoinhibition and relief mechanism by the proteolytic processing of Toll‐like receptor 8. Proc. Natl. Acad. Sci. U. S. A. 113, 3012–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanji, H. , Ohto, U. , Shibata, T. , Taoka, M. , Yamauchi, Y. , et al. (2015) Toll‐like receptor 8 senses degradation products of single‐stranded RNA. Nat. Struct. Mol. Biol. 22, 109–115. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Z. , Ohto, U. , Shibata, T. , Krayukhina, E. , Taoka, M. , et al. (2016) structural analysis reveals that toll‐like receptor 7 is a dual receptor for guanosine and single‐stranded RNA. Immunity 45, 737–748. [DOI] [PubMed] [Google Scholar]
- 20.Ito, T. , Amakawa, R. , Kaisho, T. , Hemmi, H. , Tajima, K. , et al. (2002) Interferon‐alpha and interleukin‐12 are induced differentially by Toll‐like receptor 7 ligands in human blood dendritic cell subsets. J. Exp. Med. 195, 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mancuso, G. , Gambuzza, M. , Midiri, A. , Biondo, C. , Papasergi, S. , et al. (2009) Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat. Immunol. 10, 587–594. [DOI] [PubMed] [Google Scholar]
- 22.Pandey, S. , Kawai, T. , and Akira, S. (2015) Microbial sensing by Toll‐like receptors and intracellular nucleic acid sensors. Cold Spring Harbor Perspect. Biol. 7, a016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cervantes, J. L. , Weinerman, B. , Basole, C. , and Salazar, J. C. (2012) TLR8: the forgotten relative revindicated. Cell. Mol. Immunol. 9, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlee, M. (2013) Master sensors of pathogenic RNA ‐ RIG‐I like receptors. Immunobiology 218, 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kell, A. M. , and Gale, M. Jr. (2015) RIG‐I in RNA virus recognition. Virology 479‐480, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoneyama, M. , Kikuchi, M. , Natsukawa, T. , Shinobu, N. , Imaizumi, T. , et al. (2004) The RNA helicase RIG‐I has an essential function in double‐stranded RNA‐induced innate antiviral responses. Nat. Immunol. 5, 730–737. [DOI] [PubMed] [Google Scholar]
- 27.Sumpter, R., Jr. , Loo, Y. M. , Foy, E. , Li, K. , Yoneyama, M. , et al. (2005) Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG‐I. J. Virol. 79, 2689–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sohn, J. , and Hur, S. (2016) Filament assemblies in foreign nucleic acid sensors. Curr. Opin. Struct. Biol. 37, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, B. , and Hur, S. (2015) How RIG‐I like receptors activate MAVS. Curr. Opin. Virol. 12, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vabret, N. , and Blander, J. M. (2013) Sensing microbial RNA in the cytosol. Front. Immunol. 4, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryant, C. E. , Orr, S. , Ferguson, B. , Symmons, M. F. , Boyle, J. P. , et al. (2015) International Union of Basic and Clinical Pharmacology. XCVI. Pattern recognition receptors in health and disease. Pharmacol. Rev. 67, 462–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu, Z. , and Chai, J. (2016) Structural mechanisms in nlr inflammasome assembly and signaling. Curr. Top. Microbiol. Immunol. 397, 23–42. [DOI] [PubMed] [Google Scholar]
- 33.Prochnicki T., Mangan M.S., and Latz, E. (2016) Recent insights into the molecular mechanisms of the NLRP3 inflammasome activation. F1000Research 5, 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen, I. C. , Scull, M. A. , Moore, C. B. , Holl, E. K. , McElvania‐TeKippe, E. , et al. (2009) The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30, 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanneganti, T. D. , Body‐Malapel, M. , Amer, A. , Park, J. H. , Whitfield, J. , et al. (2006) Critical role for Cryopyrin/Nalp3 in activation of caspase‐1 in response to viral infection and double‐stranded RNA. J. Biol. Chem. 281, 36560–36568. [DOI] [PubMed] [Google Scholar]
- 36.Sander, L. E. , Davis, M. J. , Boekschoten, M. V. , Amsen, D. , Dascher, C. C. , et al. (2011) Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 474, 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feerick, C. L. , and McKernan, D. P. (2016) Understanding the regulation of pattern recognition receptors in inflammatory diseases ‐ a 'Nod' in the right direction. Immunology 150, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabbah, A. , Chang, T. H. , Harnack, R. , Frohlich, V. , Tominaga, K. , et al. (2009) Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 10, 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapoor, A. , Forman, M. , and Arav‐Boger, R. (2014) Activation of nucleotide oligomerization domain 2 (NOD2) by human cytomegalovirus initiates innate immune responses and restricts virus replication. PloS One 9, e92704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hornung, V. (2014) SnapShot: nucleic acid immune sensors, part 1. Immunity 41, 868. [DOI] [PubMed] [Google Scholar]
- 41.Li, X. D. , and Chen, Z. J. (2012) Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife 1, e00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oldenburg, M. , Kruger, A. , Ferstl, R. , Kaufmann, A. , Nees, G. , et al. (2012) TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance‐forming modification. Science 337, 1111–1115. [DOI] [PubMed] [Google Scholar]
- 43.Hidmark, A. , von Saint Paul, A. , and Dalpke, A. H. (2012) Cutting edge: TLR13 is a receptor for bacterial RNA. J. Immunol. 189, 2717–2721. [DOI] [PubMed] [Google Scholar]
- 44.Sparrer, K. M. , and Gack, M. U. (2015) Intracellular detection of viral nucleic acids. Curr. Opin. Microbiol. 26, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, P. , An, H. , Liu, X. , Wen, M. , Zheng, Y. , et al. (2010) The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta‐catenin‐dependent pathway. Nat. Immunol. 11, 487–494. [DOI] [PubMed] [Google Scholar]
- 46.Ramos, H. J. , and Gale, M. Jr. (2011) RIG‐I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 1, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Negishi, H. , Yanai, H. , Nakajima, A. , Koshiba, R. , Atarashi, K. , et al. (2012) Cross‐interference of RLR and TLR signaling pathways modulates antibacterial T cell responses. Nat. Immunol. 13, 659–666. [DOI] [PubMed] [Google Scholar]
- 48.Szabo, A. , and Rajnavolgyi, E. (2013) Collaboration of Toll‐like and RIG‐I‐like receptors in human dendritic cells: tRIGgering antiviral innate immune responses. Am. J. Clin. Exp. Immunol. 2, 195–207. [PMC free article] [PubMed] [Google Scholar]
- 49.Ghosh, T. K. , Mickelson, D. J. , Solberg, J. C. , Lipson, K. E. , Inglefield, J. R. , et al. (2007) TLR‐TLR cross talk in human PBMC resulting in synergistic and antagonistic regulation of type‐1 and 2 interferons, IL‐12 and TNF‐alpha. Int. Immunopharmacol. 7, 1111–1121. [DOI] [PubMed] [Google Scholar]
- 50.Makela, S. M. , Strengell, M. , Pietila, T. E. , Osterlund, P. , and Julkunen, I. (2009) Multiple signaling pathways contribute to synergistic TLR ligand‐dependent cytokine gene expression in human monocyte‐derived macrophages and dendritic cells. J. Leukocyte Biol. 85, 664–672. [DOI] [PubMed] [Google Scholar]
- 51.Wang, J. , Shao, Y. , Bennett, T. A. , Shankar, R. A. , Wightman, P. D. , et al. (2006) The functional effects of physical interactions among Toll‐like receptors 7, 8, and 9. J. Biol. Chem. 281, 37427–37434. [DOI] [PubMed] [Google Scholar]
- 52.Demaria, O. , Pagni, P. P. , Traub, S. , de Gassart, A. , Branzk, N. , et al. (2010) TLR8 deficiency leads to autoimmunity in mice. J. Clin. Investig. 120, 3651–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarz, H. , Posselt, G. , Wurm, P. , Ulbing, M. , Duschl, A. , et al. (2013) TLR8 and NOD signaling synergistically induce the production of IL‐1beta and IL‐23 in monocyte‐derived DCs and enhance the expression of the feedback inhibitor SOCS2. Immunobiology 218, 533–542. [DOI] [PubMed] [Google Scholar]
- 54.Morosky, S. A. , Zhu, J. , Mukherjee, A. , Sarkar, S. N. , and Coyne, C. B. (2011) Retinoic acid‐induced gene‐I (RIG‐I) associates with nucleotide‐binding oligomerization domain‐2 (NOD2) to negatively regulate inflammatory signaling. J. Biol. Chem. 286, 28574–28583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1