Identification of Plasmodium falciparum Erythrocyte Membrane Protein 1 (PfEMP1) as the Rosetting Ligand of the Malaria Parasite P. falciparum (original) (raw)
Abstract
Severe Plasmodium falciparum malaria is characterized by excessive sequestration of infected and uninfected erythrocytes in the microvasculature of the affected organ. Rosetting, the adhesion of _P. falciparum_–infected erythrocytes to uninfected erythrocytes is a virulent parasite phenotype associated with the occurrence of severe malaria. Here we report on the identification by single-cell reverse transcriptase PCR and cDNA cloning of the adhesive ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1). Rosetting PfEMP1 contains clusters of glycosaminoglycan-binding motifs. A recombinant fusion protein (Duffy binding-like 1–glutathione S transferase; Duffy binding-like-1–GST) was found to adhere directly to normal erythrocytes, disrupt naturally formed rosettes, block rosette reformation, and bind to a heparin-Sepharose matrix. The adhesive interactions could be inhibited with heparan sulfate or enzymes that remove heparan sulfate from the cell surface whereas other enzymes or similar glycosaminoglycans of a like negative charge did not affect the binding. PfEMP1 is suggested to be the rosetting ligand and heparan sulfate, or a heparan sulfate–like molecule, the receptor both for PfEMP1 binding and naturally formed erythrocyte rosettes.
The mortality of human malaria is caused by Plasmodium falciparum, an intracellular protozoan that invades and multiplies in liver and red blood cells during its vertebrate life cycle. The virulence of the parasite is associated with the capacity of the infected erythrocyte to adhere to endothelial cells and to erythrocytes, which is known as rosetting. This may cause impaired local oxygen delivery and thereby death of the human host (1–3). Cerebral malaria is the most malignant form of the infection due to massive sequestration of infected and uninfected erythrocytes in the brain microvasculature.
After being transported from the internal parasite, antigens involved in the binding of cells are thought to be concentrated and subsequently exposed to the exterior of the erythrocyte at minute (∼100 nm diam), electron-dense excrescences called knobs (4). P. falciparum erythrocyte membrane protein 1 (PfEMP1) is one such antigen, a polypeptide of 200–350 kD encoded by the var family of P. falciparum genes (5–7). The feature of antigenic variation and switching of the surface of the parasitized RBC (pRBC) has been attributed to the var genes. Up to 150 such genes are harbored in the genome, but only one PfEMP1 is thought to be expressed at any one time. PfEMP1 has features of an adhesive molecule and has been associated with the cytoadherent properties of the infected red cell (8, 9). For example, the expression of PfEMP1-encoding var genes has been shown to correlate with the capacity of the pRBC for binding to host receptors, including CD36 and intercellular adhesion molecule (ICAM)-1 (9, 10).
A role for PfEMP1 in rosetting was recently suggested by Rowe et al. (11) and complement receptor 1 was found to be an associated host counterpart. However, the ABO blood group antigens, as well as CD36, were previously discovered to act as rosetting receptors (12, 13) and P. falciparum rosettes are furthermore disrupted by low concentrations of heparin, an effect that is immediate and reversible (14, 15). The latter suggested that the parasite also uses the negatively charged, sulfated glycosaminoglycans (GAG) as rosetting receptors. GAGs are composed of repeated units of sulfated or acetylated disaccharides and are present in the human body bound to a protein core in the form of proteoglycans (PGs; 16). Heparan sulfate and chondroitin sulfate are GAGs exposed on all human cell surfaces, but they are diverse and vary from cell to cell and may be within the same cell due to secondary modifications of the extensive carbohydrate chains (deacetylations, _O_-sulfations, etc.). Heparin is only found in mast cells and is characterized by a higher degree of sulfation and epimerization than heparan sulfate. However, heparan sulfate and heparin are similar due to heparin-like stretches also found in heparan sulfate (17). Polypeptides with a capacity to adhere to the negatively charged GAGs have been suggested to be composed of clusters of positively charged amino acid residues contained in the consensus sequences XBBXBX or XBBBXXBX where B is a basic residue such as lysine, arginine, or histidine (18). These motifs have been identified both in human polypeptides and in various microbial invaders and they have been discovered to mediate binding both at a linear and at a three-dimensional level (19).
Here we describe the identification by single-cell reverse transcriptase PCR (RT-PCR) of the PfEMP1 of a rosetting parasite. Clusters of GAG-binding motifs were found in the sequence. Recombinant PfEMP1 adheres to solid-phase heparin, to heparan sulfate on the erythrocyte surface, and disrupts rosettes. Further, naturally formed rosettes also seem to be mediated by binding to the same GAG.
Materials and Methods
The Parasites.
The P. falciparum parasites were cultured according to standard methods with 10% AB+ Rh+ serum added to the buffered medium (RPMI supplemented with Hepes, gentamycin, and sodium bicarbonate). The FCR3S1.2 was cloned with micromanipulation from the previously limiting-dilution cloned parasite FCR3 (20). Its rosetting rate was routinely >80% (R+). FCR3S/a was negatively enriched for low rosetting from FCR3 using Ficoll-isopaque (<10%; R−). It should be noted that FCR3S was previously called Palo Alto (Uganda) in our publications (14, 20–23). Molecular studies of the Palo Alto parasites have revealed, however, that they are identical to parasites of the FCR3 lineage (24).
Optimization of Single P. falciparum RT-PCR.
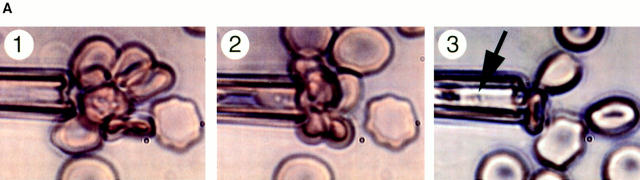
Two degenerate primers (Duffy binding-like [DBL]-1.1, 5′-GG[A/T] GC[A/T] TG[TC] GC[A/T] CC[A/T] T[A/T][T/C] [A/C]G-3′; DBL-1.2, 5′-A[A/G][A/G]T A[T/C]TG [T/A]GG [A/T]AC [A/G]TA [A/G]TC-3′) that mapped to the conserved region of all PfEMP1 DBL-1 were modified from the sequences of Su et al. The amplification parameters were first optimized so that the amplified products were visible with normal ethidium bromide staining (25). In brief, one to five parasites, obtained by limiting dilution, were directly emerged in the RT-PCR buffer (Stratagene Corp., La Jolla, CA) with different concentration of primers, MgCl2, KCl, and Tris-HCl. Both DNA and RNA were released from the parasites by heating at 93°C for 3 min. The DNA was degraded by addition of 10 U DNase (Stratagene Corp.). Reverse transcription was carried out immediately after addition of random primers and reverse transcriptase (Perkin-Elmer Corp., Norwalk, CT). The PCR reaction was subsequently performed in the same tube. Through comparison of the amplification efficiency from different reactions, the optimized parameters for single-cell RT-PCR were found to be as follows: 100 mM Tris-Cl, pH 8.3, 3.5 mM MgCl2, 500 mM KCl, and the final concentration of primers was 1 μM. In the subsequent experiments, individual trophozoite-infected rosetting erythrocytes were isolated with a 5-μm glass pipette using an inverted microscope. The selected pRBC was stripped of uninfected RBCs and repeatedly grabbed, ejected, and turned to conclusively ensure that it had pigment and that the selected cell was a single trophozoite-infected RBC (see Fig. 1 A). 50 cycles of amplification at 93°C for 20 s, 55°C for 30 s, and 72°C for 1 min were needed for product detection. Several controls were included in each experiment; one blank control (without parasite[s]) and one without reverse transcriptase to rule out the possibility of contamination and amplification due to the presence of genomic DNA.
Figure 1.
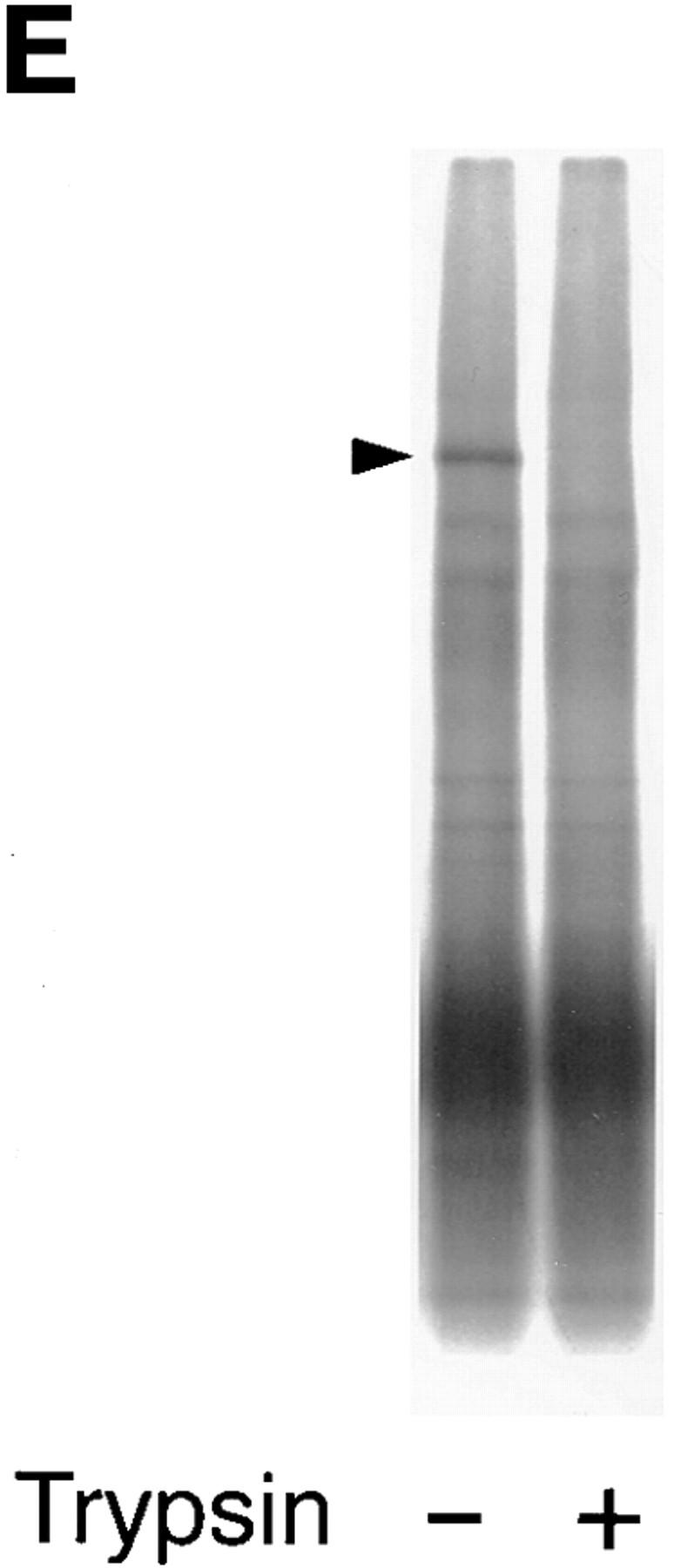
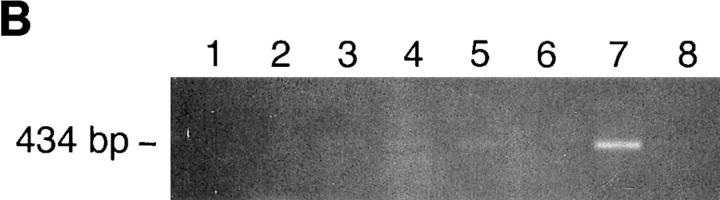
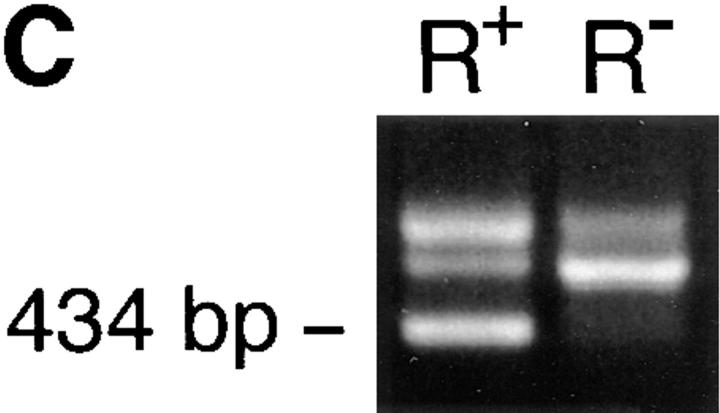
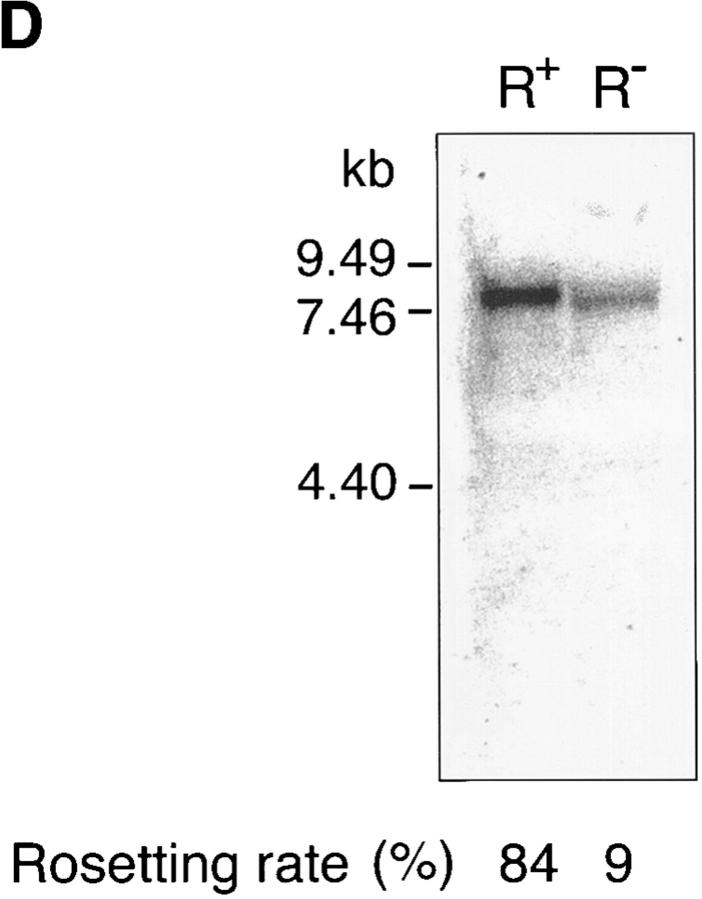
Identification of rosetting PfEMP1. (A) Rosetting, single _P. falciparum_–infected erythrocyte is seen by light microscopy held by a 5-μm micropipette (A, 1). The uninfected erythrocytes are stripped of the infected cell and careful examination confirms that it indeed is infected by a single parasite (A, 2–3). B shows the amplification of a 434-bp band in four (from reactions 3, 4, 5, and 7) out of eight single-infected, rosetting erythrocytes using degenerate primers generated from the primary sequence of the DBL-1 domain of PfEMP1. C shows the amplification pattern with the same primers as in B of bulk cultures of rosetting (R+) FCR3S1.2 cultures and the R− FCR3s/a parasites. Note that the 434-bp product is only seen with the R+ parasites. D shows the hybridization pattern in Northern blotting of the 434-bp sequence to mRNA extracted from the highly rosetting parasite FCR3S1.2 (84% R+) and the weak hybridization to the R− FCR3S/a parasite (9% R+). E shows the autoradiogarph of a Triton X-100 insoluble, SDS-soluble extract of FCR3S1.2-infected erythrocytes after radio-iodination labeling. PfEMP1 (arrow) is labeled on FCR3S1.2-infected erythrocytes and is cleaved by low concentrations of trypsin.
RT-PCR with Total RNA from Bulk-cultured FCR3S1.2 or FCR3S/a and Northern blot Analysis.
Total RNA purification from both FCR3S1.2 (R+) and FCR3S/a (R−) parasites and RT-PCR was performed as described (26). All the amplified products were cloned with the Original TA Cloning Kit (Invitrogen, Carlsbad, CA) and sequenced. Northern blot analysis was carried out using standard methods (26). Membranes were probed overnight at 60°C using the 434-bp fragment labeled with α-[32P]dCTP. Washing was performed under stringent conditions (60°C, 0.1× SSC) and the blots were examined in a phosphoimager (Molecular Dynamics, Sunnyvale, CA).
Cloning and Sequencing of the Whole cDNA.
A specific upstream primer (L-6, 5′-GAC ATG CAG CAA GGA GCT TGA TAA-3′) in the 434-bp sequence and a downstream primer (L-5, 5′-CCA TCT CTT CAT ATT CAC TTT CTG A-3′) mapping to the conserved sequence of acidic COOH-terminal segment (ATS) were generated and reverse transcription was carried out as described above. PCR was performed with the ExpandTM High Fidelity PCR System (Boehringer Mannheim GmbH, Mannheim, Germany). A single 4.9-kb fragment was amplified, which was digested into three fragments with HindIII and EcoRV and cloned into the pZErO-1 vector (Zero Background; Invitrogen). The sequencing was performed with LongRangerTM gel (FMC, Rockland, ME) on an A.L.F. Sequencer (Pharmacia, Biotech AB, Uppsala, Sweden). The 5′ region of the FCR3S1.2-_var_1 transcript was cloned by screening a cDNA library (Schlichtherle, M., unpublished data) with the 434-bp fragment as probe and seven overlapping fragments were sequenced. The 3′ terminal region was cloned by nested RT-PCR. Reverse transcription was primed with oligo-dT and PCR was performed with a specific 5′ primer (P-1, 5′-CTT TCG ACT CTA CCA TCC T-3′) upstream of transmembrane (TM) region and a 3′ primer (P-4, 5′-TTA GAT ATT CCA TAT ATC TGA TA-3′) mapping to the COOH-terminal sequence of FCR3 (_var_2) PfEMP 1. Five overlapping fragments were sequenced. 14 overlapping clones in total were sequenced in both directions to ensure that the sequence was correct and was transcribed from a single gene.
Sequence Analysis.
The DNA and amino acid sequence analysis (editing, translation, peptidesort, plot, etc.) was performed with the Genetic Computer Group (GCG, Madison, WI) (27) program. Alignment of the deduced amino acid sequence with the published PfEMP1 sequences in EMBL/GenBank/DDBJ was made to identify the sequence. The identification of potential GAG-binding motifs in FCR3S1.2-_var_1 and in the other published PfEMP1 sequences was done by searching for the presence of either of the four amino-acid motifs: KK, KR, RK, or RR first, and then manually checking each of the identified sequences for the presence of the motifs XBBXBX or XBBBXXBX where B is a basic amino acid (K, R, H) and X is a hydropathic residue. A certain degree of liberty concerning the location of the basic residues in the motifs is acceptable (18).
Expression of DBL-1 or ATS Using the pGEX-4T-1 Vector.
Both DBL-1 and ATS fragments were amplified by specific primers (Ex-1.1, 5′-ATC GAA TTC TGC AAA AAA GAT GGA AAA GGA A-3′ and D-1, 5′-GTA TTT TTT TTG TTT GTC AAA TTG-3′ for DBL-1; Ex-2, 5′-ATC GAA TTC TCT GAA AAT TTA TTC CAA A-3′ and P-4 for ATS). The amplified fragments were inserted into the EcoRI cloning site of pGEX-4T-1 downstream of the glutathione S transferase (GST) sequence. The Escherichia coli BL21 was used as the expression strain. Expression of both fusion proteins was induced with 0.1 mM isoprophyl β-d-thiogalactoside (IPTG) at 30°C for 4 h and the fusion proteins were purified on glutathione–Sepharose (Pharmacia) as described in the instructions provided by the manufacturer (GST Gene Fusion System; Pharmacia). The expression constructs were sequenced by cycle sequencing to check that the recombinant plasmids were of the expected sequences in the correct reading frames. Thrombin cleavage of the fusion proteins was performed according to a standard procedure. Western-blot analysis of DBL-1–GST and ATS–GST fusion proteins was with a biotin-labeled anti-GST mAb (clone GST-2, IgG2b; Sigma Chemical Co., St. Louis, MO) and alkaline phosphatase–avidin (Sigma Chemical Co.) to reveal the pattern of protein expression. Although the induction of expression was at a low temperature and the purification was in the presence of a cocktail of enzyme inhibitors (0.5 mM EDTA, 1 mM Pefabloc®SC [AEBSF]; Boehringer Mannheim), there was still some breakdown of the DBL-1–GST. The fusion proteins, stained by the anti-GST mAb, disappeared with thrombin treatment, leaving a 27-kD GST protein. This information together with the knowledge that the plasmids were of the expected sequences ensured that the fusion proteins were indeed the correct ones.
Heparin Binding and Blocking Assay.
The potential binding of the fusion proteins to heparin was studied by mixing either DBL-1–GST, ATS–GST fusion protein, or GST alone (20 μl, 150 μg/ml in PBS) with 20 μl of 50% heparin–Sepharose (Pharmacia) and incubating it for 5 min at room temperature in the absence of serum; binding to uncoupled Sepharose was used as control. The heparin–Sepharose and protein mixture was washed three times in large volumes of PBST buffer (PBS plus 0.05% Tween 20) before extracting bound proteins in loading buffer for 10% SDS-PAGE. Inhibition of DBL-1–GST fusion protein binding to heparin with heparin, heparan sulfate, or chondroitin sulfate (Lövens Kemiske Fabrik, Ballerup, Denmark) was also tested. 20 μl of 150 μg/ml DBL-1–GST fusion protein was mixed separately for 5 min with an equal volume of heparin, heparan sulfate, or chondroitin sulfate, titrated from 5 to 0.25 mg/ml, before the addition of Heparin–Sepharose. The inhibitory activities were checked by SDS-PAGE.
Erythrocyte Binding and Blocking Assay.
10-well immunofluorescence glass slides were precoated with 10% poly-l-lysine in PBS for 30 min. Monolayers of RBCs were made by addition of 20 μl of 0.5% 3× washed bloodgroup O Rh+RBC in PBS to each well. 20 μl DBL-1–GST, ATS–GST, or GST alone (80 μg/ml) in PBS was added to the wells for 30 min. The DBL-1–GST fusion protein was in subsequent experiments incubated in the presence of an equal volume of heparin, heparan sulfate, or chondroitin sulfate (titrated from 10 to 0.25 mg/ml) to study the inhibitory activity of each GAG. Slides were washed three times with PBS and the fusion protein binding was detected with the biotin-labeled anti-GST mAb and an ExtraAvidin®FITC conjugate (Sigma Chemical Co.). The fluorescence was assessed Optiphot-2 UV microscope with x10 ocular and an oil lens with a magnification of 100; Nikon, Tokyo, Japan).
Rosette Disruption Assay.
The assay was performed essentially as described (12). The recombinant fusion proteins (25 μl in PBS of DBL-1–GST or ATS–GST) were added to 25 μl aliquots of an ∼80% rosetting culture of FCR3S1.2 in a microtiter plate. The mixtures were incubated for 30 min at 37°C after which the rosetting rate was scored and compared to mock-treated controls after staining with acridine orange.
Enzyme Treatment of Normal RBCs.
Human bloodgroup O Rh+ erythrocytes (5% suspension in RPMI) were, before C-FDA labeling (22), incubated for 60–90 min with either heparinase III (25°C, pH 7.5; Sigma Chemical Co.), chondroitinase ABC (37°C, pH 8.0; Sigma Chemical Co.) or with clostridium perfringens neuraminidase (37°C, pH 6.0; Sigma Chemical Co.). Cells were washed three times after treatment and resuspended in complete malaria medium, with 10% serum. Control erythrocyte suspensions were mock treated, washed, and incubated as above.
Results
Identification of Rosetting PfEMP1.
A single-cell RT-PCR assay was developed using micromanipulation with the aim of identifying and isolating the PfEMP1 of a rosetting parasite. Since the PfEMP1 messages are mostly expressed at ring stage, it is always difficult to amplify cDNA from single trophozoites, but we were successful in four out of eight pRBCs studied, and when amplified, it was always a 434-bp sequence that was found (Fig. 1 B). This was confirmed by amplification of var transcripts from bulk cultures where the 434-bp amplificate was again detected unique to the rosetting P. falciparum and was not found present in nonrosetting parasites (Fig. 1 C). Both the single-cell and the RT-PCR with the total RNA from the bulk cultures were repeated several times to make sure that the results were reproducible. Further, the 434-bp product was also found present in 7 out of 12 ring stage parasites using single-cell RT-PCR (data not shown). Sequence analysis revealed that the amplified product encoded the semiconserved sequence of the 5′-located DBL-1 domain of the var genes. 10 434-bp sequences obtained from separate amplifications were found to be identical. 9 distinct PCR-amplified var transcripts from nonrosetting parasites were also isolated, subcloned, and sequenced. They were in all instances different from the 434-bp sequence (not shown) and only one of them contained a single GAG-binding motif (see below and not shown). Northern-blot analysis with messenger RNA (mRNA) from the FCR3S1.2 clone and the 434-bp sequence as the labeled probe revealed a transcript of ∼7.5 kb where the difference with the size of the coding cDNA of 6.7 kb is accounted for by a relatively large untranslated 5′ region (Sundström, A., unpublished data). The weak hybridization seen with mRNA from R− parasites is probably due to hybridyzation to a different transcript (Fig. 1 D). Thus, we conclude that a unique PfEMP1 transcript is found in rosetting parasites.
cDNA Structure of Rosetting PfEMP1.
The entire coding region of the var transcript containing the 434-bp motif was assembled with the 13 overlapping fragments and the coding sequence was found to be composed of 6,684 bp (Fig. 2 D), which encodes a 2,228 amino acid (aa) polypeptide with an estimated molecular weight of 260 kD. A single trypsin-sensitive polypeptide of a similar size was seen after 125I-lactoperoxidase surface labeling of the FCR3S1.2 (Fig. 1 E).
Figure 2.
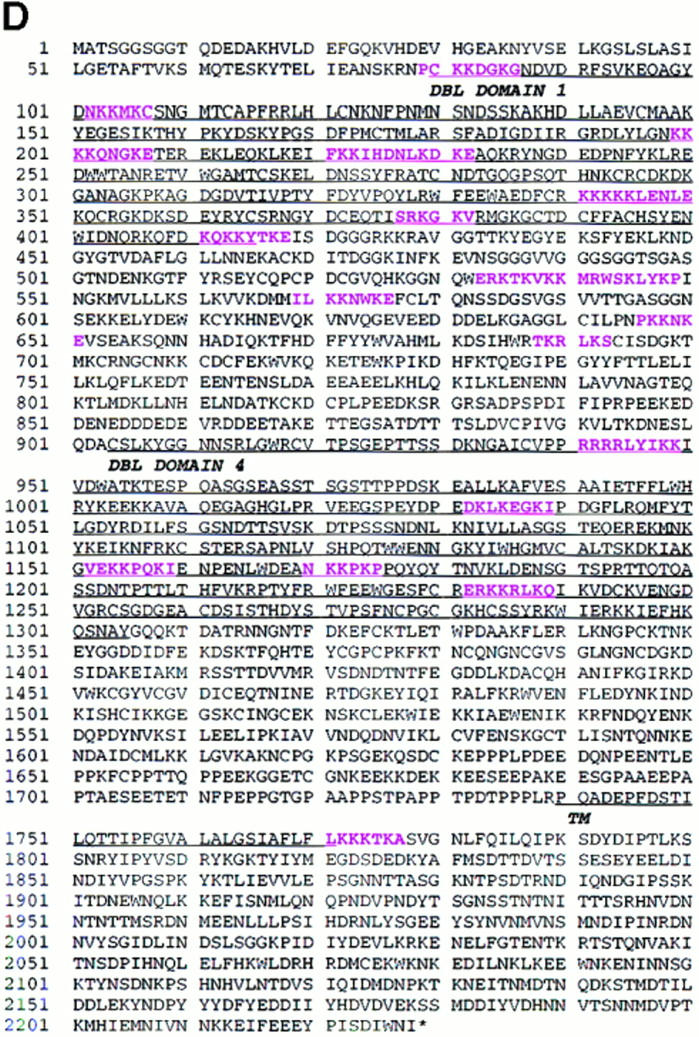
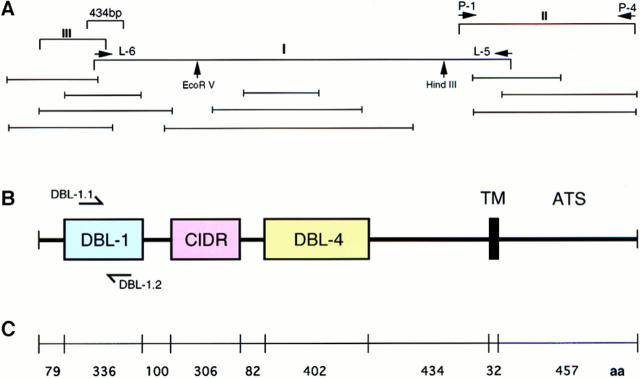
Map of cDNA structure, sequencing clones, deduced amino acid sequence, and the location of GAG-binding motifs in the rosetting PfEMP1 of FCR3S1.2. A shows the location of the 434-bp fragment and the three fragments (I, II, and III) that were initially cloned for sequencing. Restriction enzyme digestion sites are indicated by arrows. Additional overlapping clones used for sequencing are shown below. B shows the primary structure of the rosetting FCR3S1.2-PfEMP1. It has two DBL domains (DBL-1 and -4), one CIDR, one TM region, and one ATS. C shows the distribution of aa in different regions of FCR3S1.2-PfEMP1. D shows the complete aa sequence of FCR3S1.2-PfEMP1. The location of potential GAG-binding motifs are shown in pink. Motifs No. 4, 5 and 9, 10 (aa 221–232 and 533–549, respectively) are seen as a single stretch since they are located next to each other. (See Materials and Methods for description of identification of GAG-binding motifs.) These sequence data are available from EMBL/GenBank/ DDBJ under accession number AF003473.
In an in-depth analysis of the expressed FCR3S1.2-_var_1 transcript, we found that the overall structure was similar to the published var sequences. However, it was shorter than most previous sequences since it contained two DBL domains (DBL-1 and -4), rather than four, separated by a cysteine-rich interdomain region (CIDR; Fig. 2 B). Both the TM region and the negatively charged ATS of the FCR3S1.2-_var_1 were highly homologous (∼80%) to previously published sequences. However, neither the potentially variable sequences of 434-bp var gene fragment nor the other regions of the cDNA were contained in other published var sequences.
Rosetting PfEMP1 Contains Clusters of Glycosaminoglycan-binding Motifs.
We examined the FCR3S1.2-_var_1 sequence for the presence of potential GAG-binding motifs since we had previously found that rosetting was a heparin-sensitive cell-to-cell interaction. We identified 19 potential GAG-binding sequences and found that ∼95% of them (18/19) were located in the NH2 terminus. 8/19 were situated in DBL-1, 5/19 in the CIDR COOH terminus to DBL-1 and 5/19 in DBL-4. Only one potential GAG-binding motif was seen beyond the TM region (Fig. 2 D). The NH2-terminal distribution of these aa clusters is therefore consistent with the accessibility of the FCR3S1.2-_var_1 at the surface of the pRBC and may explain the molecular background to rosetting.
Rosetting PfEMP1 Binds to a Heparan Sulfate–like GAG.
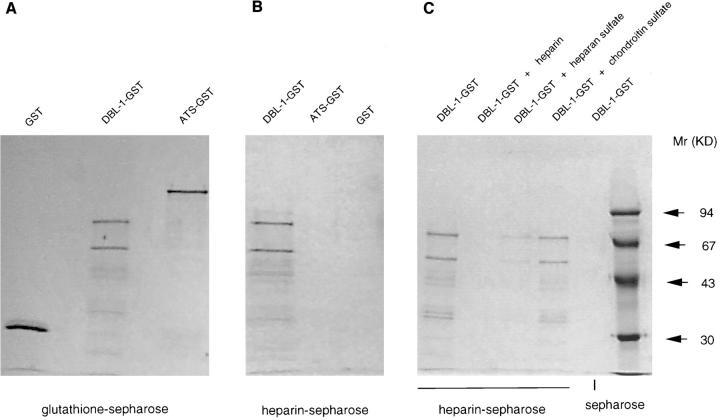
To confirm the above findings experimentally, we subsequently expressed two domains of the FCR3S1.2-_var_1 transcript: one that had eight GAG-binding motifs (DBL-1, 1,008 bp corresponding to 336 aa) and one that was the highly charged, ATS that lacks GAG-binding motifs (1,353 bp corresponding to 451 aa). The purified DBL-1–GST efficiently bound to heparin-coupled Sepharose already after a few minutes at room temperature, whereas both the ATS–GST fusion protein and a second DBL-1–GST construct covering a distinct var sequence (var 2; reference 6) that lacks GAG-binding motifs failed to bind to the heparin matrix (Fig. 3 B and not shown). The adhesion could be competed out in a dose dependent manner with heparin or heparan sulfate but not with chondroitin sulfate, another negatively charged erythrocyte surface expressed GAG (Fig. 3 C). Taken together, these indicate that the binding of DBL-1 to heparan sulfate is dependent on structure and not merely ionic interactions between the two molecules. Therefore, heparan sulfate or a heparan sulfate–like GAG seems to be the binding target for this PfEMP1.
Figure 3.
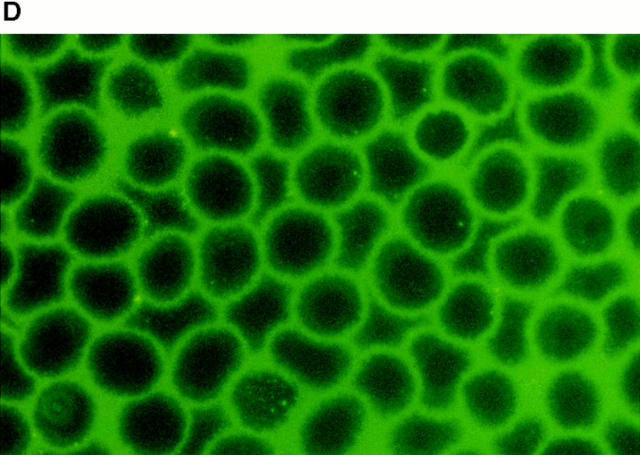
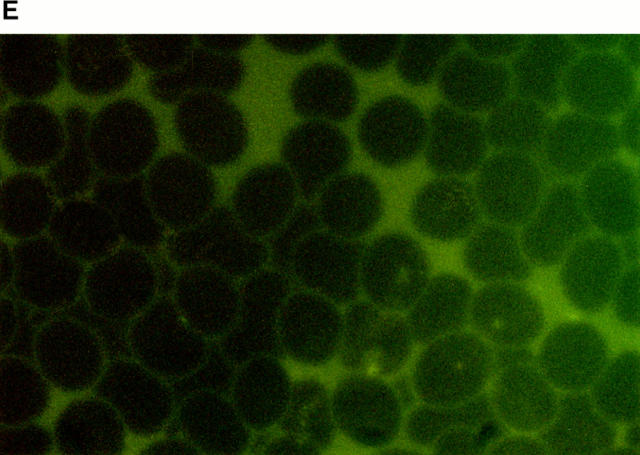
Rosetting FCR3S1.2-PfEMP1 binds to heparan sulfate. All the gels are 10% SDS-PAGE stained with Coomassie. A shows the expressed GST, DBL-1–GST, or ATS–GST after purification on glutathione–Sepharose and SDS-PAGE. B shows the binding capacity of different fusion proteins to heparin–Sepharose after SDS-PAGE. C shows the inhibition produced by different GAGs (20 μl, 5 mg/ml) on the binding of DBL-1–GST to heparin–Sepharose followed by SDS-PAGE. D and E show the binding of DBL-1–GST (D) and ATS–GST (E) to monolayers of normal RBCs as visualized by an mAb to GST labeled with biotin and FITC-avidin.
Rosetting PfEMP1 Binds Directly to Erythrocytes.
To study the potential binding of the recombinant PfEMP1 to erythrocytes we formed monolayers of uninfected erythrocytes on glass slides. The cells were subsequently incubated with different concentrations of the fusion proteins. Although the ATS–GST did not bind to the erythrocytes as detected with an anti-GST mAb, the DBL-1–GST gave a distinct surface staining of all the uninfected erythrocytes (Fig. 3, D and E). This was titratable and could be inhibited with heparin (∼50% inhibition at 1.25 mg/ml, 100% inhibition at 5 mg/ml) or heparan sulfate (∼50% inhibition at 2.5 mg/ ml, 100% inhibition at 10 mg/ml), but not with the related GAG, chondroitin sulfate, which suggests that the binding was specific. A second DBL-1–GST construct covering a distinct var sequence (_var_2; reference 6) that lacks GAG-binding motifs also failed to bind to the erythrocytes (not shown). Heparan sulfate has been previously suggested to be present on human erythrocytes (28–31) and we therefore conclude that the DBL-1–GST fusion protein binds with heparan sulfate specifically to both the solid-phase matrix as well as to normal erythrocytes.
Recombinant DBL-1 Disrupts Rosettes and Blocks Rosette Reformation.
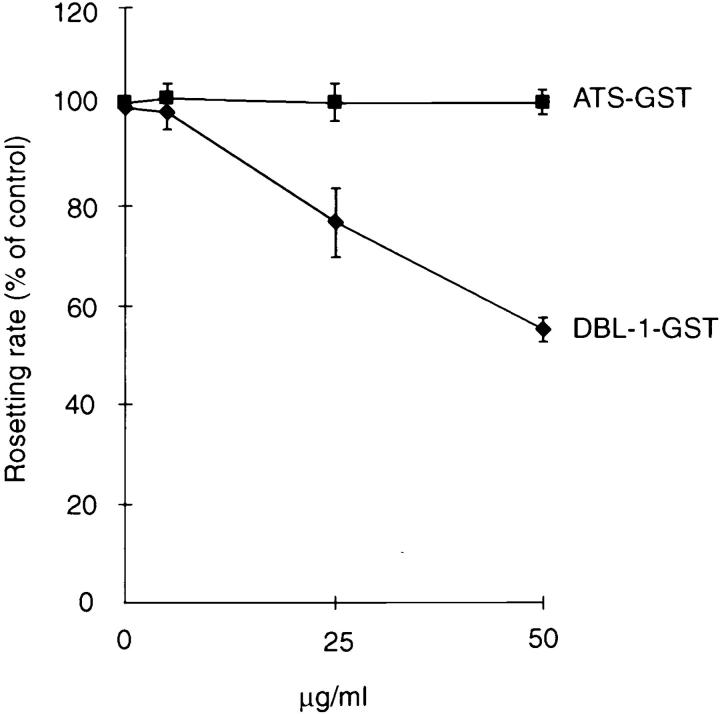
The effect of the DBL-1 fusion protein on preformed rosettes was studied to confirm the biological role of PfEMP1 in rosetting. Aliquots of a highly rosetting FCR3S1.2 culture were incubated with decreasing concentrations of DBL-1–GST, ATS–GST, or a second DBL-1–GST construct covering a distinct var sequence (_var_2; reference 6) that lacks GAG-binding motifs. DBL-1 caused a dose-dependent rosette reversion with 40–50% reversion at ∼50 μg/ml, whereas neither the ATS–GST nor the _var_2 DBL-1 showed any effect on rosettes (Fig. 4 and not shown). The rosettes were not reformed upon prolonged incubation.
Figure 4.
Disruption of preformed, natural rosettes with either DBL-1–GST or ATS–GST. Results are means and standard errors of three experiments.
Rosetting Is Dependent on a Heparan Sulfate–like GAG.
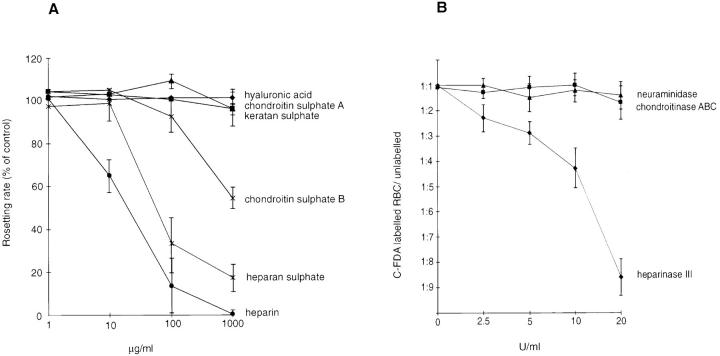
To establish the role of heparan sulfate also in rosetting, we studied the disruptive activity of different GAGs on FCR3S1.2 rosettes. FCR3S1.2 rosettes were sensitive to both heparin and to heparan sulfate, but neither chondroitin sulfate A, keratan sulfate, nor hyaluronic acid had any effect on the rosettes (Fig. 5 A). Chondroitin sulfate B had a slight effect only at high concentrations (Fig. 5 A). These findings were confirmed by enzyme treatment of the uninfected erythrocyte; heparinase, but not chondroitinase or neuraminidase, treatment blocked the rosetting (Fig. 5 B). Addition of heparan sulfate to the heparinase–cell mixture completely blocked the heparinase activity suggesting that the heparinase activity seen was indeed due to the cleavage of a heparan sulfate–like GAG and not due to a contamination causing proteolytic cleavage of erythrocyte surface–located polypeptides.
Figure 5.
Effect of GAGs on P. falciparum rosetting. A shows disruption of rosettes exerted by different GAGs. FCR3S1.2 cultures were incubated with GAGs for 1 h at 37°C and compared to control culture. Results are the means and standard error of three separate experiments. B shows the effect of enzyme treatment of uninfected, C-FDA–labeled erythrocytes in a competitive assay of rosette reformation in the presence of normal erythrocytes and FCR3S1.2-infected pRBCs. Results are the means and standard error of three separate experiments, or two experiments for neuraminidase. Neuraminidase and chondroitinase ABC concentrations are in IU, whereas the heparinase III concentration is in Sigma units. (One Sigma unit corresponds to ∼1.7 × 10−3 IU.)
Discussion
Herein we described the identification of PfEMP1 as the rosetting ligand and heparan sulfate, or a heparan sulfate–like molecule, as the rosetting receptor. A single-cell RT-PCR technique was developed to investigate the association of PfEMP1 expression with rosetting binding at the single-cell level. The PfEMP1 messages were found to be present in ring-stage parasites, in trophozoites, and in schizonts, although they are less abundant in the more mature stage parasites. However, these were studied in great detail since they do form rosettes and rings do not. One PfEMP1 mRNA species was amplified from single-rosetting parasites and the appearance of the same fragment in amplifications with total RNA from R+ bulk cultures, but not with total RNA from non- or low-rosetting parasites, indicated that the message was unique to rosetting parasites. It was evident, however, that the parasites were not homogeneous, even in cultures of a high-rosetting rate, suggesting that the nonrosetting parasites may express different PfEMP1s. Similarly, although we found strong hybridization to a ∼7.5-kb message from the rosetting parasites, there was still a small hybridization to a slightly smaller species with RNA from the R− parasites. This probably reflects the transcription of a different var gene since the FCRS1.2 _var_1 sequence was not found in the amplified products from R− parasites.
Cloning of full-length cDNA from eukaryotic cells is always difficult. To achieve this, we first optimized the RT-PCR parameters so that most PfEMP1 transcripts could be amplified. With these conditions and a primer sequence from the unique 434-bp rosetting PfEMP1 cDNA we were able to amplify one large single down-stream fragment of 4.9 kb of cDNA, which upon the digestion with EcoRI and HindIII was fragmented into distinct bands indicating that the amplified product was composed of one sequence. The 4.9-kb cDNA species was subsequently confirmed to be a single amplificate by sequencing. Rosetting PfEMP1– specific sequences of both the variable region, upstream to the trans-membrane domain, and of the 434-bp fragment were used to obtain the sequences of overlapping regions of the 3′ and the 5′ regions of this transcript. Five overlapping fragments were sequenced to ascertain the 3′ overlap and seven to ascertain the 5′ overlap. The correctness was further checked by RT-PCR with specific primers flanking the overlapping regions after the assembly of the entire coding sequence. Each of the 14 PCR-amplified fragments were sequenced in both directions and the overlapping fragments were found to be correct and identical in all regions. Therefore, it is highly likely that the assembled transcript is indeed derived from one single gene.
The high sensitivity of rosettes to heparin made us speculate that the parasite uses sulfated carbohydrates as the rosetting receptor. The appearance of potential heparin-binding motifs in this PfEMP1 sequence was therefore checked after the assembly of the transcript. Clusters of heparin-binding motifs, the consensus sequence of which has been previously proposed by others, were then identified. All the motifs showed an identical or a similar composition to other published heparin- or heparan sulfate–binding motifs found in both malaria parasites and other organisms (32, 33). The expressed DBL-1, which had eight motifs, bound to heparin–Sepharose, to the membrane of normal RBCs, disrupted naturally formed rosettes, and blocked rosette reformation while the ATS fusion protein did not. The inhibitory activity of heparan sulfate, as well as the inability of chondroitin sulfate to inhibit or block the attachment, informed us that it was the molecular structure of the GAG that is important for binding. In complementary experiments, the DBL-1 region of a second var gene transcript (_var_-2) that lacks GAG-binding motifs was expressed and found not to bind to the heparin–Sepharose. Furthermore, it did not bind to the erythrocyte surface, nor did it disrupt rosettes, which also supports a functional role of the GAG-binding sequences in the rosetting DBL-1.
The rosette reversion obtained by the addition of recombinant DBL-1 to naturally formed rosettes indicated that the parasite uses PfEMP1 as the rosetting ligand. The deletion of rosetting by heparinase treatment of normal RBCs gave us further confirmation about this specific ligand–receptor interaction indeed suggesting that heparan sulfate, or a heparan sulfate–like molecule, is involved in the binding. Heparinase treatment also disrupted the rosettes of other strains of parasites (TM180, TM284), whereas the rosettes of the strain R29 were not affected (not shown). Further, the DBL-1 domain of R29 contains only one GAG-binding motif (11). These findings, taken together with those of Rowe et al. (11), suggest that another receptor (i.e., CR1) may play a role in rosetting of R29 (11). In separate experiments, heparin-derived mono- and disaccharides that were of the same net charge but had their sulfate groups at different positions were studied for their rosette-disrupting activities. Those with the sulfate groups in the same position as heparan sulfate inhibited binding, whereas the others did not (Barragan, A., unpublished data, and see above). Furthermore, although the _N_-sulfated monosaccharide glucosamine, a component of heparan sulfate, had a good antirosetting activity, the identically _N_-sulfated galactosamine had no such activity (Barragan, A., unpublished data). Heparin has also been found to disrupt rosettes from 27 out of 54 fresh P. falciparum isolates (50%), suggesting that heparan sulfate is used by these parasites as rosetting receptors (14). The receptors used by the heparin-resistant fresh isolates may be other GAGs, as we have found that rosettes of most parasites may be disrupted by either one or the other of the human GAGs (Barragan, A., unpublished data). Thus, we conclude that FCR3S1.2 uses heparan sulfate, or a heparan sulfate–like molecule, on the erythrocyte surface as a rosetting receptor, and that the other GAGs may be receptors of other parasites.
Heparan sulfate is a molecule present on almost all cells; on erythrocytes it may be expressed on CD44 (see above and references 28–31). 4 × 106 heparan sulfate molecules have been calculated to be expressed at the surface of normal hepatocytes (34) and the density of the heparan sulfate– like molecule on the RBCs could also be high if this indeed is the receptor used by the parasite for rosetting. The involvement of GAGs as receptor structures for other cell-to-cell interactions of P. falciparum has previously been suggested, e.g., for endothelial binding, and for liver and erythrocyte invasion (32, 35–37). Taken together, these may indicate that the parasite has adapted a more general strategy for interacting with the host by using the negatively charged proteoglycans as receptors exposed on the exterior of every cell surface.
Acknowledgments
This work was supported by grants from the United Nations Development Program/World Bank/World Health Organization Special Program for Research and Training in Tropical Diseases (TDR), the Swedish Medical Research Council, and the Karolinska Institutet. Q. Chen was partially supported by the Changchun University of Agriculture and Animal Sciences, Changchun, China.
Footnotes
1
Abbreviations used in this paper: aa, amino acid; ATS, acidic COOH-terminal segment; CIDR, cysteine-rich interdomain region; DBL, Duffy binding-like; GAG, glycosaminoglycan; GST, glutathione S transferase; mRNA, messenger RNA; PfEMP 1, Plasmodium falciparum erythrocyte membrane protein 1; pRBC, parasitized RBC; RT-PCR, reverse transcriptase PCR; TM, transmembrane.
References
- 1.Miller LH, Good F, Milon G. Malaria pathogenesis. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 2.Pasloske BL, Howard RJ. Malaria, the red cell, and the endothelium. Annu Rev Med. 1994;45:283–295. doi: 10.1146/annurev.med.45.1.283. [DOI] [PubMed] [Google Scholar]
- 3.Marsh K, English M, Crawley J, Peshu N. The pathogenesis of severe malaria in African children. Ann Trop Med Parasitol. 1996;90:395–402. doi: 10.1080/00034983.1996.11813068. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson, C.T., and M. Aikawa. 1990. Ultrastructure of malaria-infected erythrocytes. Blood Cells (NY). 16:351–368. [PubMed]
- 5.Howard RJ, Barnwell JW, Kao V. Antigenic variation in Plasmodium knowlesimalaria: identification of the variant antigen on infected erythrocytes. Proc Natl Acad Sci USA. 1983;80:4129–4133. doi: 10.1073/pnas.80.13.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su X-Z, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum–infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 7.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparumgene encoding PfEMP1, a malaria variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 8.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum vargenes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baruch DI, Gormley JA, Ma C, Howard RJ, Pasloske BL. Plasmodium falciparumerythrocyte membrane protein 1 is a parasitised erythrocyte receptor for adherence to CD36, thrombospondin, and intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1996;93:3497–3502. doi: 10.1073/pnas.93.8.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner JP, Pinches RA, Roberts DJ, Newbold CI. Variant antigens and endothelial receptor adhesion in Plasmodium falciparum. . Proc Natl Acad Sci USA. 1996;93:3503–3508. doi: 10.1073/pnas.93.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparumrosetting mediated by a parasite-variant erythrocyte membrane protein and complement–receptor 1. Nature. 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- 12.Carlson J, Wahlgren M. Plasmodium falciparumerythrocytes rosetting is mediated by promiscuous lectin-like interactions. J Exp Med. 1992;176:1311–1317. doi: 10.1084/jem.176.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handunetti SM, van Schravendijk MR, Hasler T, Barnwell JW, Greenwalt DE, Howard R J. Involvement of CD36 on erythrocytes as a rosetting receptor for Plasmodium falciparum–infected erythrocytes. Blood. 1992;80:2097–2104. [PubMed] [Google Scholar]
- 14.Carlson J, Ekre HP, Helmby H, Gysin J, Greenwood BM, Wahlgren M. Disruption of Plasmodium falciparumerythrocyte rosettes by standard heparin and heparin devoid of anticoagulant activity. Am J Trop Med Hyg. 1992;46:595–602. doi: 10.4269/ajtmh.1992.46.595. [DOI] [PubMed] [Google Scholar]
- 15.Rowe A, Berendt AR, Marsh K, Newbold CI. Plasmodium falciparum: a family of sulphated glycoconjugates disrupts erythrocyte rosettes. Exp Parasitol. 1994;79:506–516. doi: 10.1006/expr.1994.1111. [DOI] [PubMed] [Google Scholar]
- 16.Yanagisha M, Hascall V. Cell surface heparan sulfate proteoglycans. J Biol Chem. 1992;267:9451–9454. [PubMed] [Google Scholar]
- 17.Faham S, Hileman ER, Fromm RJ, Lindardt JR, Rees CD. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 18.Cardin AD, Weintraub HJR. Molecular modeling of protein–glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 19.Hata A, Ridinger DN, Sutherland S, Emi M, Shuhua Z, Myers RL, Ren K, Cheng T, Inoue I, Wilson DE. Binding of lipoprotein lipase to heparin. J Biol Chem. 1993;268:8447–8457. [PubMed] [Google Scholar]
- 20.Udomsangpetch R, Wåhlin B, Carlson J, Berzins K, Torii M, Aikawa M, Perlmann P, Wahlgren M. Plasmodium falciparum–infected erythrocytes form spontaneous erythrocyte rosettes. J Exp Med. 1989;169:1835–1840. doi: 10.1084/jem.169.5.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholander C, Treutiger CJ, Hultenby K, Wahlgren M. Novel fibrillar structure confers adhesive property to malaria-infected erythrocytes. Nat Med. 1996;2:204–208. doi: 10.1038/nm0296-204. [DOI] [PubMed] [Google Scholar]
- 22.Carlson J, Holmquist G, Taylor DW, Perlmann P, Wahlgren M. Antibodies to a histidine-rich protein (PfHRP1) disrupt spontaneously formed Plasmodium falciparumerythrocyte rosettes. Proc Natl Acad Sci USA. 1990;87:2511–2515. doi: 10.1073/pnas.87.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmby H, Cavelier L, Pettersson U, Wahlgren M. Rosetting Plasmodium falciparum–infected erythrocytes express unique antigens on their surface. Infect Immun. 1993;61:284–288. doi: 10.1128/iai.61.1.284-288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fandeur T, Bonnefoy S, Mercereau-Puijalon O. In vivo and in vitro derived Palo Alto lines of Plasmodium falciparumare genetically unrelated. Mol Biochem Parasitol. 1991;47:167–178. doi: 10.1016/0166-6851(91)90176-7. [DOI] [PubMed] [Google Scholar]
- 25.Cobb BD, Clarkson JM. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res. 1994;22:3801–3805. doi: 10.1093/nar/22.18.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E.F., Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trybala E, Svennerholm B, Bergström T, Olofsson S, Jaensson S, Goodman JL. Herpes simplex virus type 1–induced hemagglutination: glycoprotein C mediates virus binding to erythrocyte surface heparan sulfate. J Virol. 1993;67:1278–1285. doi: 10.1128/jvi.67.3.1278-1285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baggio B, Marzaro G, Gambaro G, Marchini F, Williams HE, Borsatti A. Glycosaminoglycan content, oxalate self-exchange and protein phosphorylation in erythrocytes of patients with ‘idiopathic' calcium oxalate nephrolithiasis. Clin Sci. 1990;79:113–116. doi: 10.1042/cs0790113. [DOI] [PubMed] [Google Scholar]
- 30.Anstee DJ, Gardner B, Spring FA, Holmes CH, Simpson KL, Parsons SF, Mallinson G, Yousaf SM, Judson PA. New monoclonal antibodies in CD44 and CD58: their use to quantify CD44 and CD58 on normal erythrocytes and to compare the distribution of CD44 and CD58 in human tissues. Immunology. 1991;74:197–205. [PMC free article] [PubMed] [Google Scholar]
- 31.Elenius K, Jalkanen M. Function of the syndecans: a family of cell surface proteoglycans. J Cell Sci. 1994;107:2975–2982. doi: 10.1242/jcs.107.11.2975. [DOI] [PubMed] [Google Scholar]
- 32.Sinnis P, Clavijo P, Fenyö D, Chait BT, Cerami C, Nussenzweig V. Structural and functional properties of region II–plus of the malaria circumsporozoite protein. J Exp Med. 1994;180:297–306. doi: 10.1084/jem.180.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson LR, Busch JS, Cardin DA. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol Rev. 1991;71:481–530. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- 34.Kjellén L, Oldberg Å, Höök M. Cell surface heparan sulfate. J Biol Chem. 1980;21:10407–10413. [PubMed] [Google Scholar]
- 35.Kulane A, Ekre H-P, Perlmann P, Rombo L, Wahlgren M, Wåhlin B. Effect of different fractions of heparin on Plasmodium falciparum merozoite invasion of red blood cells in vitro. . Am J Trop Med Hyg. 1992;46:589–594. doi: 10.4269/ajtmh.1992.46.589. [DOI] [PubMed] [Google Scholar]
- 36.Robert C, Pouvelle B, Meyer P, Muanza K, Scerf A, Gysin J. Chondroitin-4-sulfate (proteoglycan) as Plasmodium falciparum–infected erythrocyte adherence receptor of brain microvascular endothelial cells. Res Immunol. 1995;146:383–393. doi: 10.1016/0923-2494(96)81042-x. [DOI] [PubMed] [Google Scholar]
- 37.Rogerson SJ, Chaiyaroj SC, Ng K, Reeder JC, Brown GV. Chondritin sulfate A is a cell surface receptor for Plasmodium falciparum–infected erythrocytes. J Exp Med. 1995;182:15–20. doi: 10.1084/jem.182.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]