Characterization of Staphylococci with Reduced Susceptibilities to Vancomycin and Other Glycopeptides (original) (raw)
Abstract
During the last several years a series of staphylococcal isolates that demonstrated reduced susceptibility to vancomycin or other glycopeptides have been reported. We selected 12 isolates of staphylococci for which the vancomycin MICs were ≥4 μg/ml or for which the teicoplanin MICs were ≥8 μg/ml and 24 control strains for which the vancomycin MICs were ≤2 μg/ml or for which the teicoplanin MICs were ≤4 μg/ml to determine the ability of commercial susceptibility testing procedures and vancomycin agar screening methods to detect isolates with reduced glycopeptide susceptibility. By PCR analysis, none of the isolates with decreased glycopeptide susceptibility contained known vancomycin resistance genes. Broth microdilution tests held a full 24 h were best at detecting strains with reduced glycopeptide susceptibility. Disk diffusion did not differentiate the strains inhibited by 8 μg of vancomycin per ml from more susceptible isolates. Most of the isolates with reduced glycopeptide susceptibility were recognized by MicroScan conventional panels and Etest vancomycin strips. Sensititre panels read visually were more variable, although with some of the panels MICs of 8 μg/ml were noted for these isolates. Vitek results were 4 μg/ml for all strains for which the vancomycin MICs were ≥4 μg/ml. Vancomycin MICs on Rapid MicroScan panels were not predictive, giving MICs of either ≤2 or ≥16 μg/ml for these isolates. Commercial brain heart infusion vancomycin agar screening plates containing 6 μg of vancomycin per ml consistently differentiated those strains inhibited by 8 μg/ml from more susceptible strains. Vancomycin-containing media prepared in-house showed occasional growth of susceptible strains, Staphylococcus aureus ATCC 29213, and on occasion, Enterococcus faecalis ATCC 29212. Thus, strains of staphylococci with reduced susceptibility to glycopeptides, such as vancomycin, are best detected in the laboratory by nonautomated quantitative tests incubated for a full 24 h. Furthermore, it appears that commercial vancomycin agar screening plates can be used to detect these isolates.
Vancomycin and teicoplanin are glycopeptides with significant activity against gram-positive bacterial pathogens (5, 30). Although teicoplanin has not been approved for use in the United States, vancomycin is widely used for the treatment of infections caused by staphylococci and enterococci (7, 22, 24, 30).
In 1988, reports of high-level vancomycin resistance in enterococci (vancomycin-resistant enterococci [VRE]) caused significant concern among the medical community (20, 37). Concern increased when it was discovered that many of the isolates of VRE in the United States also proved to be resistant to beta-lactams and aminoglycosides, leaving few therapeutic options (6, 18, 22, 24). VRE are also a growing concern in hospitals in Europe (1, 15). In 1992, Noble and colleagues (28) showed that the vanA operon could be transferred by cell-to-cell matings between enterococci and staphylococci; however, clinical isolates of staphylococci containing the vanA, vanB, vanC, or vanD gene have not been reported.
Recently, Hiramatsu and colleagues (17) reported the first clinical isolate of Staphylococcus aureus with reduced susceptibility to vancomycin. Subsequently, similar organisms with reduced susceptibility to glycopeptides were reported from Michigan (9) and New Jersey (9). All three organisms were methicillin-resistant strains of S. aureus that had decreased susceptibility to vancomycin in vivo after prolonged exposure to that drug (8, 9, 17).
In this report, we characterize these three isolates of S. aureus and nine other staphylococci with various degrees of susceptibility to glycopeptides. These isolates are discussed by using the acronym GISA (glycopeptide-intermediate Staphylococcus aureus) or GISS (glycopeptide-intermediate Staphylococcus species) since many of these isolates are also resistant to teicoplanin, a glycopeptide commonly used in Europe and other parts of the world. For convenience, glycopeptide-susceptible isolates of S. aureus and Staphylococcus species will be referred to as GSSA and GSSS, respectively.
MATERIALS AND METHODS
Bacterial isolates.
The bacterial isolates with reduced glycopeptide susceptibility used in this study are described in Table 1. These include eight isolates submitted to the Centers for Disease Control and Prevention from seven states and four S. aureus isolates from Japan. Strains Mu50 and Mu3 were isolated at Juntendo University Hospital, Tokyo, Japan. Mu3-8R was derived from Mu3 by selection on vancomycin-containing agar (16). N20 was isolated from a patient at Nagasaki University Hospital, Nagasaki, Japan. These isolates have been described elsewhere (16). S. aureus 963sm and 966 were obtained from a patient in Michigan courtesy of Barbara Robinson-Dunn, Michigan Department of Public Health, and isolate 992, from the blood of a patient in New Jersey, was provided by Sandra Pine, Our Lady of Lourdes Medical Center, Camden, N.J. Both isolates have been described elsewhere (9). One isolate of S. aureus from a patient in Florida (isolate 803) was provided by Tim Sellen (Jacksonville, Fla.). Three isolates of Staphylococcus epidermidis, i.e., 5289, 12333, and 759, were provided by Judith Johnston (West Sacramento, Calif.), Jane Wong (San Francisco, Calif.), and Carol Spiegel (Madison, Wis.), respectively. Isolate 5289 was described previously (14). The Staphylococcus haemolyticus isolate (isolate 142) was provided by Ken Van Horn (Valhalla, N.Y.). An additional 24 glycopeptide-susceptible isolates from the United States were included as controls: 9 methicillin-resistant S. aureus, 7 methicillin-susceptible S. aureus, and 6 S. epidermidis isolates selected from the culture collection of the Centers for Disease Control and Prevention and 2 S. aureus isolates from the American Type Culture Collection (ATCC). Organisms were identified by standard biochemical procedures (19).
TABLE 1.
Comparison of vancomycin and teicoplanin MICs and disk diffusion zone sizes with the interpretive results for isolates of staphylococci exhibiting decreased susceptibility to glycopeptides
| Isolate (source) | Vancomycin | Teicoplanin | ||
|---|---|---|---|---|
| MICa | DDb | MIC | DD | |
| S. aureus Mu50 (Japan) | 8 (I) | 17 (S) | 8 (S) | 15 (S) |
| S. aureus Mu3 (Japan) | 2 (S) | 19 (S) | 16 (I) | 14 (S) |
| S. aureus Mu3-8R (Japan) | 8 (I) | 17 (S) | 32 (R) | 13 (I) |
| S. aureus N20 (Japan) | 4 (S) | 18 (S) | 1 (S) | 17 (S) |
| S. aureus 963sm (Michigan) | 8 (I) | 18 (S) | 16 (I) | 13 (I) |
| S. aureus 966 (Michigan) | 4 (S) | 16 (S) | 16 (I) | 13 (I) |
| S. aureus 992 (New Jersey) | 8 (I) | 17 (S) | 8 (S) | 15 (S) |
| S. aureus 803 (Florida) | 4 (S) | 17 (S) | 16 (I) | 15 (S) |
| S. epidermidis 5289 (Virginia) | 8 (I) | 18 (S) | 16 (I) | 14 (S) |
| S. epidermidis 759 (Wisconsin) | 8 (I) | 17 (S) | 16 (I) | 14 (S) |
| S. epidermidis 12333 (California) | 8 (I) | 17 (S) | 16 (I) | 13 (I) |
| S. haemolyticus 142 (New York) | 8 (I) | 17 (S) | 32 (R) | 13 (I) |
Amplification of vanA, vanB, and vanC genes by PCR.
The oligonucleotide primers and the reaction conditions previously reported for the amplification of vanA, vanB, vanC1, vanC2, and vanC3 in enterococci were used (11, 12, 31). A Perkin-Elmer Cetus DNA thermocycler (Applied Biosystems, Foster City, Calif.) was programmed with the following conditions: initial denaturation, 10 min at 95°C; 30 cycles with a 30-s denaturation step at 94°C, a 30-s annealing step at 58°C, and a 30-s extension step at 72°C; a 10-min extension step at 72°C; and a holding step at 4°C until the sample was analyzed. Samples of the PCR products were electrophoresed, stained with 10 μM ethidium bromide, and visualized and photographed by using UV transillumination. The control strains of enterococci used in this study included E. faecalis A256 (vanA), E. faecalis V583 (vanB), E. gallinarum NJ4 (vanC1), E. casseliflavus ATCC 25788 (vanC2), and E. flavescens ATCC 49996 (vanC3). Isolates were not tested for the presence of the vanD gene.
PFGE and arbitrarily primed PCR typing.
The protocol for the preparation of _Sma_I- and _Apa_I-digested chromosomal DNA for pulsed-field gel electrophoresis (PFGE) analysis was that described by Bannerman et al. (4). For electrophoresis, the plugs were cut into small slices (2 by 5 mm) and placed into 125 μl of a total restriction enzyme mixture (restriction buffer plus sterile distilled water) containing 20 U of _Sma_I or _Apa_I (New England Biolabs, Beverly, Mass.). After a 2-h incubation at 25°C with shaking at 140 rpm, chromosomal restriction fragments were separated by loading the trimmed slices of the plug into a well of 1% SeaKem agarose running gel (FMC Corp., Rockland, Maine). The running gel was prepared in 0.5× TBE buffer (Bio-Rad, Richmond, Calif.). The wells containing the plugs were sealed with 1% SeaPlaque agarose. Electrophoresis was performed with a CHEF-DR III electrophoresis cell (Bio-Rad, Melville, N.Y.). Bacteriophage lambda DNA concatemers (Bio-Rad) were used as size standards and served as a control for the running parameters of the CHEF-DR III unit. The running parameters were as follows: initial pulse, 5 s; final pulse, 40 s; voltage, 6 V/cm; time, 20 h; temperature, 12 to 14°C. The gels were stained with ethidium bromide and photographed.
The banding patterns were interpreted visually by published guidelines (36). Dice coefficients were generated with Advanced Quantifier 1-D Match software, version 2.5 (Bio Image, Ann Abor, Mich.), with a 3.0% band tolerance setting. Arbitrarily primed PCR was performed with Ready-To-Go RAPD Analysis beads (Pharmacia Biotech) by using the primer 5′-CTA GGA CCG C-3′ according to the manufacturer’s directions.
Antimicrobial susceptibility testing.
Isolates were tested by the broth microdilution method described by the National Committee for Clinical Laboratory Standards (NCCLS) in document M7-A4 (27) with cation-adjusted Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.). The antimicrobial agents tested included chloramphenicol, ciprofloxacin, clindamycin, erythromycin, gentamicin, oxacillin, penicillin, rifampin, tetracycline, trimethoprim-sulfamethoxazole, and vancomycin. Five investigational antimicrobial agents were included in the broth microdilution tests; teicoplanin (Hoechst Marion Roussel, Cincinnati, Ohio), LY333328 (Eli Lilly, Indianapolis, Ind.), dalfopristin plus quinupristin (Rhone-Poulenc Rohrer, Collegeville, Pa.), linezolid (Pharmacia-Upjohn, Kalamazoo, Mich), and SCH27899 (Schering-Plough, Kenilworth, N.J.). The presence of β-lactamase was assayed by a chromogenic nitrocefin assay (29). All isolates were tested for susceptibility to vancomycin and teicoplanin by the NCCLS reference disk diffusion method as described in document M2-A6 (26) by using 30-μg vancomycin and teicoplanin disks.
Vancomycin MICs were determined for each isolate by using five commercial systems; bioMérieux Vitek (Hazelwood, Mo.) GPS-101 cards (version R05.01), MicroScan conventional panels (Combo 6; Dade Behring Inc., MicroScan Division, West Sacramento, Calif.) read on the MicroScan Walk/Away (DMS version 20.3), MicroScan Rapid Pos Combo 1 panels read on the MicroScan Walk/Away, AccuMed International, Inc. (Westlake, Ohio) Sensititre MD panels read visually, and the Etest (AB Biodisk North America, Inc., Piscataway, N.J.). All systems and methods were performed according to the respective manufacturers’ instructions. Mueller-Hinton agar (Becton Dickinson Microbiology Systems [BDMS], Cockeysville, Md.) was used for Etest studies.
Vancomycin agar screen.
In-house-prepared brain heart infusion agar (BHIA) screening plates with vancomycin and base BHIA medium from Acumedia Manufacturers, Inc. (Baltimore, Md.), BDMS, Difco Laboratories (Detroit, Mich.), Oxoid, Inc. (Ogdensburg, N.Y.), and Remel (Lenexa, Kans.), each containing either 2, 4, or 6 μg of vancomycin per ml, were evaluated by testing the 12 GISA and GISS isolates and the 24 GSSA and GSSS isolates. Commercially prepared BHIA screening plates were evaluated by testing all 36 isolates on one lot each of BHIA medium from BDMS, Hardy Diagnostics (Santa Maria, Calif.), PML Microbiologicals (Wilsonville, Oreg.), and Remel. All plates were inoculated by preparing, in sterile water, a suspension from growth from a 20- to 24-h blood agar plate to a turbidity equivalent to that of a 0.5 McFarland standard, dropping 10 μl of the inoculum onto the agar surface with a calibrated pipettor, and incubating the plates at 35°C for 24 h (34, 35).
Quality control.
Quality control of commercial products was performed by using the organisms recommended by the respective manufacturer. S. aureus ATCC 29213 and E. faecalis ATCC 29212 were used for quality control of broth microdilution tests, S. aureus ATCC 25923 was used for quality control of disk diffusion tests, and E. faecalis ATCC 29212 and E. faecalis ATCC 51299 were used for quality control of the vancomycin screening plates. S. aureus ATCC 29213 and S. aureus ATCC 25923 were also used to evaluate the vancomycin screening plates.
RESULTS
Reference susceptibility tests and analysis of isolates by PCR.
Broth microdilution and disk diffusion susceptibility tests were performed with 36 isolates of staphylococci which included 12 GISA or GISS isolates for which the vancomycin MICs were ≥4 μg/ml or for which the teicoplanin MICs were ≥8 μg/ml (8 isolates of S. aureus, 3 isolates of S. epidermidis, and 1 isolate of S. haemolyticus) and 24 GSSA or GSSS isolates for which the vancomycin MICs were ≤2 μg/ml or for which the teicoplanin MICs were ≤4 μg/ml. The susceptibility test results for GISA and GISS isolates are presented in Table 1. For four of the S. aureus isolates, i.e., for Mu50 and Mu3-8R (a derivative of Mu3 that was selected on vancomycin-containing agar), 963sm, and 992, the vancomycin MICs were repeatedly 8 μg/ml by the broth microdilution method (NCCLS interpretation, intermediate), but the zones of inhibition were 17 or 18 mm (NCCLS interpretation, susceptible) by disk diffusion testing. For the three isolates of S. epidermidis, i.e., 5289, 759, and 12333, and S. haemolyticus 142, the MICs were also 8 μg/ml (intermediate) but the disk zone sizes were 17 or 18 mm (susceptible). For S. aureus 803, 966, and N20, the vancomycin MICs were 4 μg/ml and the zone sizes were 16 to 18 mm. For one isolate, Mu3, the vancomycin MIC was 2 μg/ml and the zone size was 19 mm. For all GSSA and GSSS isolates vancomycin disk zone sizes were ≥16 mm.
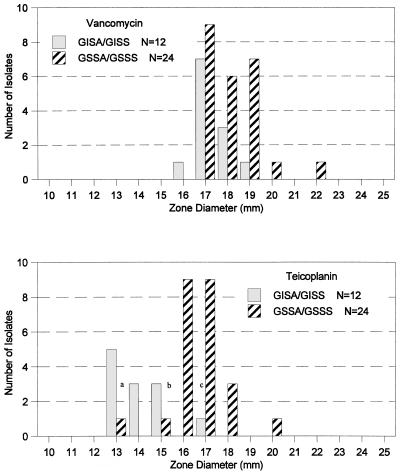
Teicoplanin MICs were also elevated by the broth microdilution reference method for the 11 isolates for which vancomycin MICs were ≥4 μg/ml. For isolate Mu3 (vancomycin MIC, 2 μg/ml), the teicoplanin MIC was 16 μg/ml (intermediate). For all eight isolates inhibited by 8 μg of vancomycin per ml, the teicoplanin MICs were 8 to 32 μg/ml. Of these isolates, for two strains the teicoplanin MICs were 32 μg/ml (resistant), for four strains the teicoplanin MICs were 16 μg/ml (intermediate), and for the two remaining strains the teicoplanin MICs were 8 μg/ml (susceptible). For isolates 966 and 803, inhibited by 4 μg of vancomycin per ml by the reference method, the teicoplanin MICs were 16 μg/ml. The remaining isolate for which the vancomycin MIC was 4 μg/ml, S. aureus N20, was inhibited by a much lower concentration of teicoplanin, 1 μg/ml. The teicoplanin disk diffusion zone sizes for the two isolates for which the teicoplanin MICs were 32 μg/ml were 13 mm, while the seven isolates for which the teicoplanin MICs were 16 μg/ml had zone sizes of 13 to 15 mm. The two isolates for which the teicoplanin MICs were 8 μg/ml had zone sizes of 15 mm. Two control isolates, an S. haemolyticus isolate and an S. epidermidis isolate, shown in Fig. 1 as “a” and “b,” were inhibited by 4 μg of teicoplanin per ml and zone diameters were 13 and 15 mm, respectively. For the remaining 22 control isolates the teicoplanin MICs were ≤2 μg/ml and the disk diffusion zone sizes were ≥16 mm. Figure 1 illustrates the correlation between disk diffusion zone sizes and MICs for vancomycin and teicoplanin.
FIG. 1.
Distribution of disk diffusion zone diameters of Staphylococcus species obtained with a 30-μg vancomycin disk or a 30-μg teicoplanin disk. Discrepant results are (a) an S. epidermidis (GSSS) isolate for which the teicoplanin zone diameter was 13 mm and the teicoplanin MIC was 4 μg/ml, (b) an S. epidermidis (GSSS) isolate for which the teicoplanin zone diameter was 15 mm and the teicoplanin MIC was 4 μg/ml, and (c) an S. aureus (GISA) isolate (isolate N20) for which the teicoplanin zone diameter was 17 mm and the teicoplanin MIC was 1 μg/ml.
The susceptibility patterns of the 12 GISA and GISS isolates to other antimicrobial agents determined by the broth microdilution reference method are presented in Table 2. All of the isolates, regardless of species, were resistant to clindamycin, erythromycin, oxacillin, and penicillin. The S. aureus isolates remained susceptible to trimethoprim-sulfamethoxazole. All 12 GISA and GISS isolates were negative when tested for the vanA, vanB, vanC1, vanC2, and vanC3 genes by PCR. The isolates were not tested for the presence of the vanD gene.
TABLE 2.
Resistance patterns of staphylococcal study isolates to other commonly tested antimicrobial agents determined by the broth microdilution reference methoda
| Isolate (source) | Agents to which the isolate is resistant or intermediate | Agent(s) to which the isolate is susceptible |
|---|---|---|
| S. aureus Mu50 (Japan) | P, Ox, Cd, E, Gm, Cip, Rif, T | SXT, C |
| S. aureus Mu3 (Japan) | P, Ox, Cd, E, Gm, Cip, C (I), T | SXT, Rif |
| S. aureus Mu3-8R (Japan) | P, Ox, Cd, E, Gm, Cip, C, T | SXT, Rif |
| S. aureus N20 (Japan) | P, Ox, Cd, E, Gm, Cip, C (I), T | SXT, Rif |
| S. aureus 963sm (Michigan) | P, Ox, Cd, E, Gm, Cip | SXT, C, Rif, T |
| S. aureus 966 (Michigan) | P, Ox, Cd, E, Gm, Cip, C (I) | SXT, Rif, T |
| S. aureus 992 (New Jersey) | P, Ox, Cd, E, Cip, Rif | SXT, Gm, C, T |
| S. aureus 803 (Florida) | P, Ox, Cd, E, Cip, Rif | SXT, Gm, C, T |
| S. epidermidis 5289 (Virginia) | P, Ox, Cd, E, Gm (I), Cip, SXT | C, Rif, T |
| S. epidermidis 759 (Wisconsin) | P, Ox, Cd, E, Gm, Cip, C (I), SXT | Rif, T |
| S. epidermidis 12333 (California) | P, Ox, Cd, E, Gm | SXT, C, Rif, T, Cip |
| S. haemolyticus 142 (New York) | P, Ox, Cd, E, Gm, Cip, C, SXT, T | Rif |
Susceptibility to experimental agents.
The susceptibilities of the 12 GISA and GISS isolates to four experimental antimicrobial agents were determined by the broth microdilution method. The results for the experimental agents LY333328 (a glycopeptide), dalfopristin-quinupristin (streptogramins), linezolid (an oxazolidinone also known as U100766), and SCH27899 (an everninomycin) are presented in Table 3. No interpretive criteria exist for these experimental antimicrobial agents; however, the MICs for all of the isolates were within the published MIC50s (MICs at which 50% of isolates are inhibited) or MIC90s of dalfopristin-quinupristin (3, 25) but up to 2 dilutions higher than the reported MIC90s of LY333328 (32) and linezolid (25). The MICs of SCH27899 were within the MIC90 of this drug for 100 staphylococcal isolates tested previously by our laboratory (MIC90 = 2.0 μg/ml; data not shown). All quality control results were within the ranges specified by the pharmaceutical manufacturers of each compound.
TABLE 3.
MICs of investigational antimicrobial agents for the staphylococcal isolates studied
| Isolate (source) | Vancomycin | MIC (μg/ml)a | ||||
|---|---|---|---|---|---|---|
| Teicoplanin | LY333328 | Dalfopristin-quinupristin | Linezolid | SCH27899 | ||
| S. aureus Mu50 (Japan) | 8 | 8 | 4 | 0.5 | 2 | 0.5 |
| S. aureus Mu3 (Japan) | 2 | 16 | 4 | 1 | 2 | 0.5 |
| S. aureus Mu3-8R (Japan) | 8 | 32 | 2 | 0.5 | 2 | 0.5 |
| S. aureus N20 (Japan) | 4 | 1 | 2 | 0.5 | 2 | 0.25 |
| S. aureus 963sm (Michigan) | 8 | 16 | 8 | 1 | 2 | 1 |
| S. aureus 966 (Michigan) | 4 | 16 | 4 | 1 | 2 | 0.5 |
| S. aureus 992 (New Jersey) | 8 | 8 | 4 | 0.5 | 1 | 0.25 |
| S. aureus 803 (Florida) | 4 | 16 | 8 | 0.5 | 2 | 0.25 |
| S. epidermidis 5289 (Virginia) | 8 | 16 | 8 | ≤0.25 | 2 | 0.25 |
| S. epidermidis 759 (Wisconsin) | 8 | 16 | 4 | ≤0.25 | 2 | 1 |
| S. epidermidis 12333 (California) | 8 | 16 | 4 | ≤0.25 | 2 | 0.5 |
| S. haemolyticus 142 (New York) | 8 | 32 | 8 | 0.5 | 2 | 0.5 |
Commercial susceptibility testing methods.
The 12 GISA and GISS isolates were tested for susceptibility to vancomycin by five commercial methods: Vitek, MicroScan conventional panels, MicroScan rapid panels, Sensititre panels, and Etest. The results are presented in Table 4. The MicroScan conventional panels and Etest proved to be the most effective in detecting GISA and GISS isolates. With the MicroScan conventional panels the MICs were 8 μg/ml for six of the eight isolates for which the reference MICs were 8 μg/ml and the MICs were 4 μg/ml for the other two isolates (Mu3-8R and 992). With the MicroScan conventional panels the MICs were 8 μg/ml for isolates 966 and 803, for which the reference MICs were 4 μg/ml. Etest MICs were 8 μg/ml for five of eight isolates inhibited by 8 μg/ml by the reference method, 6 μg/ml for two other isolates, and 4 μg/ml for the remaining isolate (Mu3-8R). The Etest produced an MIC of 8 μg/ml for one isolate for which the reference MIC was 4 μg/ml, isolate 966. According to the Etest package insert (2), an MIC falling between 2 doubling dilutions is interpreted as the next higher full dilution for purposes of susceptibility category interpretation. By application of their rounding criteria, the Etest interpretation for all eight isolates inhibited by 8 μg/ml was intermediate. With the Sensititre panels the MICs were 8 μg/ml for four of eight isolates for which the reference MICs were 8 μg/ml and 4 μg/ml for three isolates. Mu3-8R, the remaining isolate which was inhibited by 8 μg/ml by the reference method, was inhibited by only 2 μg/ml on the Sensititre panel. Isolate 803 was also inhibited by 8 μg/ml, although the reference MIC was 4 μg/ml. With the Vitek system the MICs were 4 μg/ml for all eight isolates for which the reference MICs were 8 μg/ml and 1 μg/ml for isolate N20, for which the reference MIC was 4 μg/ml. With the MicroScan Rapid panels the MICs were either ≥16 or ≤2 μg/ml for all isolates for which the reference MIC was 8 μg/ml; consequently, use of this method is not recommended. Isolates 966 and 803 were inhibited by 4 μg/ml by the reference method, but the MICs were reported as 8 μg/ml for each isolate with two systems: with MicroScan conventional and Etest for 966 and with MicroScan conventional and Sensititre for 803. For the GSSA and GSSS isolates the vancomycin MICs were ≤2 μg/ml by all of the above methods.
TABLE 4.
Performance of commercial antimicrobial susceptibility test systems with staphylococcal study isolates
| Isolate (source) | Vancomycin MIC (μg/ml [interpretive category]) with the following commercial susceptibility testing systema: | |||||
|---|---|---|---|---|---|---|
| Broth microdilutionb | Vitek | MicroScan conventional | MicroScan rapid | Sensititre | Etest | |
| S. aureus Mu50 (Japan) | 8 (I) | 4 (S) | 8 (I) | >16 (R) | 4 (S) | 6 (I) |
| S. aureus Mu3 (Japan) | 2 (S) | 1 (S) | ≤2 (S) | ≤2 (S) | 1 (S) | 2 (S) |
| S. aureus Mu3-8R (Japan) | 8 (I) | 4 (S) | 4 (S) | ≤2 (S) | 2 (S) | 4 (S) |
| S. aureus N20 (Japan) | 4 (S) | 1 (S) | 4 (S) | ≤2 (S) | 1 (S) | 2 (S) |
| S. aureus 963sm (Michigan) | 8 (I) | 4 (S) | 8 (I) | >16 (R) | 8 (I) | 8 (I) |
| S. aureus 966 (Michigan) | 4 (S) | 4 (S) | 8 (I) | 4 (S) | 4 (S) | 8 (I) |
| S. aureus 992 (New Jersey) | 8 (I) | 4 (S) | 4 (S) | ≤2 (S) | 8 (I) | 6 (I) |
| S. aureus 803 (Florida) | 4 (S) | 4 (S) | 8 (I) | >16 (R) | 8 (I) | 4 (S) |
| S. epidermidis 5289 (Virginia) | 8 (I) | 4 (S) | 8 (I) | >16 (R) | 8 (I) | 8 (I) |
| S. epidermidis 759 (Wisconsin) | 8 (I) | 4 (S) | 8 (I) | >16 (R) | 4 (S) | 8 (I) |
| S. epidermidis 12333 (California) | 8 (I) | 4 (S) | 8 (I) | >16 (R) | 4 (S) | 8 (I) |
| S. haemolyticus 142 (New York) | 8 (I) | 4 (S) | 8 (I) | >16 (R) | 8 (I) | 8 (I) |
Vancomycin agar screen.
Table 5 presents the results of tests with the 12 GISA and GISS isolates and 24 glycopeptide-susceptible isolates on commercial BHIA vancomycin screening plates. All eight of the isolates for which the reference vancomycin MICs were 8 μg/ml grew on the plates. All 25 isolates for which the vancomycin MICs were ≤2 μg/ml, including Mu3, were inhibited. Of the three isolates for which the reference vancomycin MICs were 4 μg/ml, one isolate, S. aureus 966, grew on all of the commercial screening plates. This isolate was obtained from the same patient in Michigan at the same time that isolate 963sm was obtained. Isolate 966 has an antibiogram profile very similar to that of 963sm, is identical to 963sm by PFGE analysis, and is vancomycin intermediate with two commercial test systems. Thus, its ability to grow on the screening plates was not unexpected.
TABLE 5.
Results of commercial vancomycin screening plates tested with staphylococcal isolates
| Isolate description | No. of plates positive/total no. testeda | |||
|---|---|---|---|---|
| BBL | Hardy | PML | Remel | |
| Vancomycin MIC, 8 μg/ml | 8/8 | 8/8 | 8/8 | 8/8 |
| Vancomycin MIC, 4 μg/ml | 1/3b | 1/3b | 1/3b | 1/3b |
| Vancomycin MIC, ≤2 μg/ml | 0/25c | 0/25 | 0/25 | 0/25 |
| Enterococcus faecalis ATCC 51299d | 5/5 | 5/5 | 5/5 | 5/5 |
| Enterococcus faecalis ATCC 29212d | 0/5 | 0/5 | 0/5 | 0/5 |
| Staphylococcus aureus ATCC 29213d | 0/5 | 0/5 | 0/5 | 0/5 |
| Staphylococcus aureus ATCC 25923d | 0/5 | 0/5 | 0/5 | 0/5 |
The results obtained with the in-house-prepared BHIA vancomycin screening plates varied with the lot of medium used. The lots of BHIA used were selected at random and were not those specifically selected by the medium manufacturers for use in the vancomycin screening assay; i.e., the lots were not prequalified. The results obtained with these lots of BHIA are presented in Table 6. Plates containing 6 μg of vancomycin per ml tended to differentiate isolates for which the MICs were 8 μg/ml from more susceptible isolates, except that three of five lots failed to detect one isolate for which the MIC was 8 μg/ml (S. epidermidis 759). Four of five lots also grew vancomycin-susceptible isolates. Isolate 966 grew on all lots of medium with vancomycin at 6 μg/ml, as it did on the commercial screening plates. Reduction of the vancomycin concentration to 4 or 2 μg/ml improved the detection of isolates for which the MICs were 8 μg/ml, but extensive growth of the susceptible isolates and quality control strains occurred.
TABLE 6.
Results of testing with vancomycin screening agar prepared in-house
| Screening plate content and isolate description | No. of plates positive/total no. tested with mediuma: | ||||
|---|---|---|---|---|---|
| Acumedia | BBL | Difco | Oxoid | Remel | |
| Vancomycin, 6 μg/ml | |||||
| Vancomycin MIC = 8 μg/ml | 7/8 | 7/8 | 8/8 | 8/8 | 7/8 |
| Vancomycin MIC = 4 μg/ml | 1/3b | 2/3b | 1/3b | 1/3b | 1/3b |
| Vancomycin MIC ≤ 2 μg/ml | 0/25c | 1/25 | 1/25 | 2/25 | 2/25 |
| E. faecalis ATCC 51299d | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| E. faecalis ATCC 29212d | 0/3 | 1/3 | 0/3 | 0/3 | 1/3 |
| S. aureus ATCC 29213d | 0/3 | 3/3 | 0/3 | 2/3 | 1/3 |
| S. aureus ATCC 25923d | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| Vancomycin, 4 μg/ml | |||||
| Vancomycin MIC = 8 μg/ml | 8/8 | 7/8 | 8/8 | 8/8 | 8/8 |
| Vancomycin MIC = 4 μg/ml | 1/3 | 2/3 | 1/3 | 1/3 | 1/3 |
| Vancomycin MIC ≤ 2 μg/ml | 3/25 | 7/25 | 2/25 | 3/25 | 13/25 |
| E. faecalis ATCC 51299 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| E. faecalis ATCC 29212 | 0/3 | 0/3 | 0/3 | 1/3 | 3/3 |
| S. aureus ATCC 29213 | 2/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| S. aureus ATCC 25923 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| Vancomycin, 2 μg/ml | |||||
| Vancomycin MIC = 8 μg/ml | 8/8 | 8/8 | 8/8 | 8/8 | 8/8 |
| Vancomycin MIC = 4 μg/ml | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| Vancomycin MIC ≤ 2 μg/ml | 13/25 | 24/25 | 21/25 | 22/25 | 24/25 |
| E. faecalis ATCC 51299 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| E. faecalis ATCC 29212 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| S. aureus ATCC 29213 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| S. aureus ATCC 25923 | 1/3 | 2/3 | 2/3 | 3/3 | 3/3 |
Tables 5 and 6 indicate that E. faecalis ATCC 51299 and E. faecalis ATCC 29212 are suitable quality control isolates. However, S. aureus ATCC 29213 appears to be a more rigorous control of test performance and may be preferred over E. faecalis ATCC 29212 for use as a susceptible quality control isolate, since it was completely inhibited on plates containing vancomycin at 6 μg/ml but grew on some lots of the in-house-prepared BHIA plates containing vancomycin at 6 μg/ml.
PFGE and arbitrarily primed PCR typing.
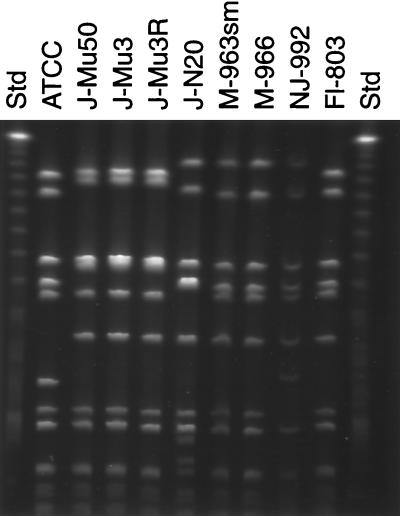
_Sma_I- or _Apa_I-digested genomic DNA from the eight GISA isolates was analyzed by PFGE to determine whether the Japanese and U.S. isolates were derived from a single clone of methicillin-resistant S. aureus (MRSA). S. aureus ATCC 29213 was included as a non-GISA control. The results of PFGE of the _Sma_I-digested DNA are presented in Fig. 2. Analysis of _Sma_I data demonstrates that Japanese isolates Mu50, Mu3, and Mu3-8R (lanes 3 to 5, respectively) are indistinguishable, while isolate N20 (lane 6) is distinctly different from the other Japanese isolates (≤70% related by the Dice coefficient with a 3.0% band tolerance; greater than six band differences). S. aureus 963sm and 966 (lanes 7 and 8, respectively), both from Michigan, are also indistinguishable from one another and are related to New Jersey 992 (lane 9) (Dice coefficient, ∼90%; two band differences) but are not related to the Japanese isolates. The Florida S. aureus isolate (lane 10) is more closely related to the three Japanese isolates (Dice coefficient, ∼90%) than to the Michigan and New Jersey isolates (Dice coefficients, <80 and <70%, respectively), although by visual inspection it appears to be much more closely related to the Michigan isolates (due to shifts in size of the two largest bands). Analysis of the _Apa_I digestion products obtained by PFGE showed that all of the isolates, including those from Japan, were highly related (data not shown). Thus, the _Sma_I profiles are more discriminatory. Arbitrarily primed PCR testing with a single decanucleotide primer did not distinguish among any of the S. aureus isolates.
FIG. 2.
PFGE patterns of _Sma_I-digested DNA from S. aureus isolates exhibiting reduced susceptibility to vancomycin. ATCC, S. aureus ATCC 29213; Std, molecular size standard.
DISCUSSION
The susceptibility of staphylococci to vancomycin and other glycopeptides appears to be waning (16). Isolates of S. aureus from Japan (8, 16, 17), Michigan (9), and New Jersey (9) and isolates of S. epidermidis (14) and S. haemolyticus from several U.S. cities were inhibited by 8 μg of vancomycin per ml, while additional isolates of S. aureus with similar PFGE profiles were inhibited by 2 to 4 μg/ml. The decrease in vancomycin susceptibility is accompanied, in most cases, by decreased susceptibility to another glycopeptide, teicoplanin, prompting us to use the terms GISA and GISS (for glycopeptide) rather than VISA (vancomycin-intermediate S. aureus). Decreased staphylococcal susceptibility to vancomycin is not due to transfer of van genes from VRE or to small colony variants, as noted in staphylococci for other antimicrobial agents (21), but appears to be a gradual selection process due to treatment pressure. Glycopeptide-resistant mutants of S. aureus have been experimentally selected by increasing the levels of vancomycin present during in vitro growth (13, 33). The GISA isolates described here may represent mutants selected in vivo with increased resistance to glycopeptides as a result of prolonged exposure of the organisms to constant levels of vancomycin in an opportune environment. Although there is speculation about the role of penicillin-binding proteins that may compete with vancomycin for binding to cellular targets and changes in cell wall turnover (23, 33), the mechanism of glycopeptide resistance remains unclear. PFGE analysis showed that three Japanese isolates (Mu50, Mu3, and Mu3-8R) were indistinguishable, as were the two Michigan isolates, but both sets were different from the fourth Japanese isolate (N20) and from each other. Thus, the isolates from Japan, Michigan, and New Jersey do not represent a single clone of MRSA that has been disseminated. By using commercial software and the criteria of a Dice coefficient of ≥85% and fewer than three band differences (36), the New Jersey isolate appeared to be related to the Michigan isolates, while the Florida isolate appeared by the same criteria to be related to the Japanese strains, but by visual comparison the Florida isolate seemed to be more closely related to the Michigan isolates. Use of alternate enzymes, such as _Apa_I, did not clarify the relationships among the strains. Arbitrarily primed DNA typing showed that all GISA strains were indistinguishable (data not shown).
Of greatest concern is that disk diffusion testing, which is widely used in the United States and around the world (38), does not differentiate strains with reduced susceptibility to vancomycin from susceptible strains (MIC range, 0.5 to 2 μg/ml). Therefore, the disk diffusion test should not be used for testing staphylococci with vancomycin. However, our data suggest that disk diffusion tests with teicoplanin may be of value for identifying isolates with reduced susceptibility to glycopeptides, although it may be necessary to modify the interpretive criteria for staphylococci to make the test more sensitive. All GSSA and GSSS isolates for which the teicoplanin MICs were ≤2 μg/ml had disk diffusion zone sizes of 16 mm or larger, and all GISA and GISS isolates for which the teicoplanin MICs were ≥8 μg/ml had disk diffusion zone sizes that were 15 mm or smaller. However, it is not clear at this point where to place strains with borderline vancomycin MICs (4 μg/ml). S. aureus N20, for example, had zone sizes of 17 mm with teicoplanin and the isolate would have been classified as susceptible, while for two susceptible GSSS isolates (one S. haemolyticus and one S. epidermidis; vancomycin and teicoplanin MICs, 4 μg/ml) teicoplanin zone sizes were 13 and 15 mm, respectively, and these isolates would have been classified as nonsusceptible by the teicoplanin disk method. Thus, the 30-μg teicoplanin disk test appears to be able to distinguish between GISA-GISS and GSSA-GSSS populations; zone sizes are ≥16 mm for the glycopeptide-susceptible organisms and ≤15 mm for the glycopeptide-nonsusceptible organisms; however, a multilaboratory study is needed to validate this test. A conservative approach may be to use the 30-μg teicoplanin disk as a screening test for decreased susceptibility to glycopeptides and to confirm the result by a broth microdilution test incubated for a full 24 h for any isolates for which the teicoplanin zone size is ≤15 mm.
Isolates with reduced vancomycin susceptibility were recognized by standard broth microdilution panels, MicroScan conventional susceptibility panels, Etest, and, to a lesser extent, Sensititre panels. With the Vitek system the MICs were consistently 4 μg/ml for GISA and GISS isolates. Inspection of the raw data generated with the Vitek system indicated that the isolates were growing in test cards but were not meeting the minimum growth criteria to be assigned higher MICs. Rapid MicroScan panels did not successfully recognize these isolates, identifying them as either completely susceptible (≤2 μg/ml) or completely resistant (>16 μg/ml). Of the automated methods, MicroScan conventional panels appear to produce results closest to that of the NCCLS broth microdilution reference method. The Etest consistently produced MIC readings within 0.5 to 1 dilution of the reference method, although the endpoint was often indistinct due to feathery growth at the edge of the ellipse. Such endpoints with coagulase-negative staphylococci and glycopeptides are recognized and discussed in the Etest package insert and may in fact be an indicator that investigators should suspect decreased glycopeptide susceptibility.
It appears that commercially prepared BHIA screening plates containing 6 μg of vancomycin per ml can be used to screen for strains of staphylococci with reduced glycopeptide susceptibility. However, agar screening plates prepared in-house showed lot-dependent growth of the susceptible E. faecalis ATCC 29212 and S. aureus ATCC 29213 control strains. Discussions with manufacturers of media confirm that lots of BHIA are screened and selected on the basis of their performance prior to being used to prepare commercial screening plates. Thus, laboratories that prepare their own media must use tight quality control for the plates, preferably with both of the susceptible control strains E. faecalis ATCC 29212 and S. aureus ATCC 29213, before using them for screening. The one S. aureus isolate, isolate 966, for which the vancomycin MIC was 4 μg/ml was obtained from the same patient infected with strain 963sm, for which the MIC was 8 μg/ml; the strains had identical PFGE profiles. Thus, it is not surprising that 966 grew on the screening plates. The BHIA screening test can be very helpful for the screening of isolates in pure culture, but since it does not contain other antimicrobial agents that inhibit the growth of other organisms, it should not be used for the direct plating of specimens.
This group of 12 isolates exhibited susceptibility characteristics that distinguished them from the control isolates. It is noteworthy that those isolates for which the vancomycin MICs were 4 μg/ml or for which the teicoplanin MICs were 8 μg/ml appear to fit the characteristics of the GISA and GISS group more closely than those of the GSSA and GSSS group and most likely are GISA or GISS isolates for which the critical MIC breakpoint has not been reached. Staphylococci, especially methicillin-resistant isolates for which the vancomycin MICs are ≥4 μg/ml or for which the teicoplanin MICs are ≥8 μg/ml, should be considered to have reduced susceptibility to glycopeptides. Under these circumstances, commercial systems, with the exception of the MicroScan rapid panels, would provide acceptable performance for the detection of staphylococci with decreased susceptibility to glycopeptides.
We recommend reading the vancomycin MICs and screening plates after a full 24 h of incubation, as is done for oxacillin and methicillin. The vancomycin MICs for any isolates growing on the screening plates should be confirmed by a broth dilution method with an incubation period of 24 h. Methicillin- or oxacillin-resistant isolates for which the vancomycin MICs are ≥4 μg/ml should be considered as having the potential for decreased susceptibility to vancomycin. In summary, glycopeptide resistance in staphylococci is becoming more common, and laboratories should be prepared to detect such strains and work with infection control personnel to contain potential outbreaks (10).
ACKNOWLEDGMENTS
We thank Theresa Smith and William Jarvis, who conducted the epidemiologic investigations with the U.S. isolates, for helpful discussions.
REFERENCES
- 1.Aarestrup F M, Ahrens P, Madsen M, Pallesen L V, Poulsen R L, Westh H. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob Agents Chemother. 1996;40:1938–1940. doi: 10.1128/aac.40.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AB Biodisk America Inc. Etest package insert. Piscataway, N.J: AB Biodisk North America Inc.; 1996. [Google Scholar]
- 3.Archer G L, Auger P, Doern G V, Ferraro M J, Fuchs P C, Jorgensen J H, Low D E, Murray P R, Reller L B, Stratton C W, Wennersten C B, Moellering R C., Jr RP 59500, a new streptogramin highly active against recent isolates of North American staphylococci. Diagn Microbiol Infect Dis. 1993;16:223–226. doi: 10.1016/0732-8893(93)90113-l. [DOI] [PubMed] [Google Scholar]
- 4.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barna J C, Williams D H. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Recommendations for preventing the spread of vancomycin resistance: recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC) Morbid Mortal Weekly Rep Rec Rep. 1995;44(12):1–20. [Google Scholar]
- 8.Centers for Disease Control and Prevention. Reduced susceptibility of Staphylococcus aureus to vancomycin—Japan, 1996. Morbid Mortal Weekly Rep. 1997;46:624–626. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morbid Mortal Weekly Rep. 1997;46:765–766. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Interim guidelines for prevention and control of staphylococcal infection associated with reduced susceptibility to vancomycin. Morbid Mortal Weekly Rep. 1997;46:626–628. , 635–636. [PubMed] [Google Scholar]
- 11.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark N C, Teixeira L M, Facklam R R, Tenover F C. Program and abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Detection and differentiation of the vanC-1, vanC-2, and vanC-3 glycopeptide resistance genes in enterococci, abstr. E-125; p. 86. [Google Scholar]
- 13.Daum R S, Gupta S, Sabbagh R, Milewski W M. Characterization of Staphylococcus aureus isolates with decreased susceptibility to vancomycin and teicoplanin: isolation and purification of a constitutively produced protein associated with decreased susceptibility. J Infect Dis. 1992;166:1066–1072. doi: 10.1093/infdis/166.5.1066. [DOI] [PubMed] [Google Scholar]
- 14.Garrett D O, Jochimsen E, Murfitt K, Hill B, McAllister S, Nelson P, Spera R, Sall R, Tenover F, Jarvis W. Abstracts of the 7th Annual Meeting of the Society for Healthcare Epidemiology of America. Infect. Control Hosp. Epidemiol. 18(Suppl.):P32. 1997. The impending apocalypse; the emergence of vancomycin resistance in Staphylococcus spp., abstr. S1. [Google Scholar]
- 15.Gordts B, Van Landuyt H, Ieven M, Vandamme P, Goossens H. Vancomycin-resistant enterococci colonizing the intestinal tracts of colonized patients. J Clin Microbiol. 1995;33:2842–2846. doi: 10.1128/jcm.33.11.2842-2846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fuckuchi Y, Kobayashi I. Vancomycin-resistant Staphylococcus aureus: dissemination of heterogeneously resistant strains in Japanese hospital. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 17.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 18.Jones R N, Sader H S, Erwin M E, Anterson S C the Enterococcus Study Group. Emerging multiply resistant enterococci among clinical isolates. I. Prevalence data from a 97 medical center surveillance study in the United States. Diagn Microbiol Infect Dis. 1995;21:85–93. doi: 10.1016/0732-8893(94)00147-o. [DOI] [PubMed] [Google Scholar]
- 19.Kloos W E, Bannerman T L. Staphylococcus and Micrococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 282–298. [Google Scholar]
- 20.Leclerq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated vancomycin and teicoplanin resistance in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 21.Mitsuyama J, Yamada H, Maehana J, Fukuda Y, Kurose S, Minami S, Todo Y, Watanabe Y, Narita H. Characteristics of quinolone-induced small colony variants in Staphylococcus aureus. J Antimicrob Chemother. 1997;39:697–705. doi: 10.1093/jac/39.6.697. [DOI] [PubMed] [Google Scholar]
- 22.Montecalvo M A, Horowicz H, Gedris C, Carbonaro C, Tenover F C, Issah A, Cook P, Wormser G P. Outbreak of vancomycin-, ampicillin-, and aminoglycoside-resistant Enterococcus faecium bacteremia in an adult oncology unit. Antimicrob Agents Chemother. 1994;38:1363–1367. doi: 10.1128/aac.38.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreira B, Boule-Vavra S, Boudewijn L, Dejonge M, Daum R S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agent Chemother. 1997;41:1788–1793. doi: 10.1128/aac.41.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris J G, Jr, Shay D K, Hebden J N, McCarter R J, Perdue B E, Jarvis W, Johnson J A, Dowling T C, Polish L B, Schwalbe R S. Enterococci resistant to multiple antimicrobial agents, including vancomycin. Ann Intern Med. 1995;123:250–259. doi: 10.7326/0003-4819-123-4-199508150-00002. [DOI] [PubMed] [Google Scholar]
- 25.Mulazimoglu L, Drenning S D, Yu V L. In vitro activities of two novel oxazolidinones (U100592 and U100766), a new fluoroquinolone (trovafloxacin), and dalfopristin-quinupristin against Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1996;40:2428–2430. doi: 10.1128/aac.40.10.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests, 6th ed., vol. 17, no. 1. Approved standard M2-A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed., vol. 17, no. 2. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 28.Noble W C, Virani Z, Cree R. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;93:195–198. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 29.O’Callaghan C H, Morris A, Kirby S M, Shingler A H. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds P E. Structure, biochemistry, and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989;8:943–950. doi: 10.1007/BF01967563. [DOI] [PubMed] [Google Scholar]
- 31.Satake S N, Clark N C, Rimland D, Nolte F S, Tenover F C. Detection of vancomycin-resistant enterococci in fecal samples by PCR. J Clin Microbiol. 1997;35:2325–2330. doi: 10.1128/jcm.35.9.2325-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwalbe R S, McIntosh A C, Qaiyumi S, Johnson J A, Johnson R J, Furness K M, Holloway W J, Steele-Moore L. In vitro activity of LY333328, an investigational glycopeptide antibiotic, against enterococci and staphylococci. Antimicrob Agents Chemother. 1996;40:2416–2419. doi: 10.1128/aac.40.10.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sieradzki K, Tomasz A. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J Bacteriol. 1997;179:2557–2566. doi: 10.1128/jb.179.8.2557-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swenson J M, Clark N C, Sahm D F, Ferraro M J, Doern G, Hindler J, Jorgensen J H, Pfaller M A, Reller L B, Weinstein M P, Zabransky R J, Tenover F C. Molecular characterization and multilaboratory evaluation of Enterococcus faecalis ATCC 51299 for quality control of screening tests for vancomycin and high-level aminoglycoside resistance in enterococci. J Clin Microbiol. 1995;33:3019–3021. doi: 10.1128/jcm.33.11.3019-3021.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swenson J M, Clark N C, Ferraro M J, Sahm D F, Doern G, Pfaller M A, Reller L B, Weinstein M P, Zabransky R J, Tenover F C. Development of a standardized screening method for detection of vancomycin-resistant enterococci. J Clin Microbiol. 1994;32:1700–1704. doi: 10.1128/jcm.32.7.1700-1704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2339. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uttley A H, Collins C H, Naidoo J, George R C. Vancomycin-resistant enterococci. Lancet. 1988;i:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 38.Verbist, L. 1993. Relevance of antibiotic susceptibility testing for clinical practice. Eur. J. Clin. Microbiol. Infect. Dis. 12(Suppl. 1)**:**2–5. [DOI] [PubMed]