Mutations in Salmonella Pathogenicity Island 2 (SPI2) Genes Affecting Transcription of SPI1 Genes and Resistance to Antimicrobial Agents (original) (raw)
Abstract
The Salmonella typhimurium genome contains two pathogenicity islands (SPI) with genes encoding type III secretion systems for virulence proteins. SPI1 is required for the penetration of the epithelial layer of the intestine. SPI2 is important for the subsequent proliferation of bacteria in the spleens of infected hosts. Although most mutations in SPI2 lead to a strong reduction of virulence, they have different effects in vitro, with some mutants having significantly increased sensitivity to gentamicin and the antibacterial peptide polymyxin B. Previously we showed that certain mutations in SPI2 affect the ability of S. typhimurium to secrete SPI1 effector proteins and to invade cultured eukaryotic cells. In this study, we show that these SPI2 mutations affect the expression of the SPI1 invasion genes. Analysis of reporter fusions to various SPI1 genes reveals highly reduced expression of sipC, prgK, and hilA, the transcriptional activator of SPI1 genes. These observations indicate that the expression of one type III secretion system can be influenced dramatically by mutations in genes encoding a second type III secretion system in the same cell.
A large number of virulence factors are important for the systemic pathogenesis of mice by Salmonella typhimurium. One group of genes important for the invasion of host epithelial cells by S. typhimurium is clustered at 63 centisomes on the chromosome (26). This virulence locus was termed Salmonella pathogenicity island 1 (SPI1), and molecular analysis of SPI1 has revealed that more than 28 genes encoding a complex type III secretion system (spa, inv, prg, and org), as well as secreted effector proteins (sip or ssp; spt) and regulatory components (invF and hilA), are involved in the invasion phenotype and induction of morphological changes of the host cell (for a review, see reference 9). Many of these invasion genes were identified by using cell culture-based screens (10, 22). By screening mutants directly in the animal host via signature-tagged mutagenesis (STM), we identified a second cluster of genes encoding a separate type III secretion system at 30 centisomes of the chromosome. This locus was termed Salmonella pathogenicity island 2 (SPI2) (16, 30). Inactivation of SPI2 genes resulted in a dramatic attenuation of virulence and the inability of mutants to colonize the spleens of infected animals. SPI2 contains genes for a type III secretion system apparatus (ssa) (17, 28, 30) and a two-component regulatory system (ssr) (28, 30), as well as candidate genes for a set of secreted effectors (sse) and their specific chaperones (ssc) (our unpublished observations). By comparison of the genetic organizations and sequences of SPI1 and SPI2 genes, it seems clear that the two systems were acquired independently from an unknown source rather than by duplication of one of the loci (15, 17).
Several phenotypic effects of SPI2 mutations have been identified. Some SPI2 mutants show reduced levels of serum resistance, whereas others appear to be more resistant (17). Some mutant strains show reduced survival inside J774 macrophages (28). Surprisingly, several SPI2 mutants are less able to invade cultured epithelial cells or macrophages, and reduced amounts of the SPI1 secreted protein SipC were detected in the culture medium (17).
These observations prompted us to undertake a more thorough analysis of SPI2 mutant phenotypes in vitro and to investigate the basis of the effect of SPI2 mutations on SPI1 secretion in more detail. In this study, we demonstrate that mutations in SPI2 affect the expression of SPI1 genes. Furthermore, mutations in SPI2 result in an altered resistance of S. typhimurium to various antibacterial agents.
MATERIALS AND METHODS
Bacterial strains, plasmids, and construction of reporter fusion strains.
S. typhimurium NCTC 12023 (identical to ATCC 14028s) was used as the wild-type strain throughout this study. All mutants and reporter fusions described were constructed in the S. typhimurium 12023 background. S. typhimurium mutations in SPI1 and SPI2 were identified after STM with a derivative of mTn_5_ as described previously (16, 27, 28a). mTn_5lacZY_ insertions in various SPI1 genes resulting in lacZ reporter gene fusions have been described previously (2, 3, 18) and were kindly provided by C. A. Lee (Harvard University). lacZ fusions were transferred into various SPI1 and SPI2 backgrounds by P22 transduction according to standard procedures (23). Lack of lysogeny of transductants was routinely confirmed by streaking on green plates (23), and correct integration of the reporter fusions was checked by Southern hybridization analysis. Details of strains used in this study are presented in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | See supplier’s documentation | Gibco BRL |
| M15[pREP] | See supplier’s documentation | Qiagen |
| S17-1 λ_pir_ | Tpr Smr_recA thi pro_ (r− m+) | |
| RP4:2-Tc:Mu:Km Tn_7 λpir_ | 8 | |
| S. typhimurium | ||
| P2D6 | ssaV::mTn_5_ (SPI2) | 30 |
| P8G12 | ssaB::mTn_5_ (SPI2)a | 30 |
| P9B6 | ssaV::mTn_5_ (SPI2) | 30 |
| P9B7 | ssaT::mTn_5_ (SPI2) | 30 |
| P11C3 | ssaV::mTn_5_ (SPI2) | 30 |
| P11D10 | ssaJ::mTn_5_ (SPI2) | 30 |
| P4H2 | hilA::mTn_5_ (SPI1) | 30 |
| P6E11 | spaRS::mTn_5_ (SPI1) | Lab stock |
| P7B12 | sipC::mTn_5_ (SPI1) | Lab stock |
| EE638 | sipC::Tn_5lacZY_11-6 (SPI1) | 18 |
| EE656 | prgH::Tn_5lacZY_ (SPI1) | 2 |
| EE658 | hilA::Tn_5lacZY_ (SPI1) | 2 |
| EE659 | prgK::Tn_5lacZY_ (SPI1) | 2 |
| NP_ssaV_ | ssaV::aphT | This study |
| Plasmids | ||
| pACYC184 | Tetr Cmr, low copy number | 6 |
| pGP704 | Ampr, R6K ori | 25 |
| pQE30 | Ampr | Qiagen |
| pJD10 | sipC in pQE30, Ampr | This study |
| pNP_ssaV_ | Kanr Ampr R6K ori | This study |
| p_ssaT_ | Cmr, ssaT in pACYC184 | This study |
| p_ssaTU_ | Cmr, ssaTU in pACYC184 | This study |
| pVV214 | HilA+ | 2 |
| pVV135 | HilA+ constitutive | 3 |
Bacterial growth conditions.
For detection of SipC in culture supernatants and total cells, as well as for reporter gene assays, bacterial cultures were grown in LB containing 1% (wt/vol) NaCl without addition of antibiotics. Cultures were grown overnight at 37°C with a culture volume of 3.5 ml in glass test tubes with agitation of 50 rpm in a roller drum. Proteins secreted by S. typhimurium were precipitated as described previously (18). Briefly, bacteria were pelleted by ultracentrifugation for 20 min at 125,000 × g at 4°C. Residual cellular debris was removed by filtration through 0.45-μM-pore-size nitrocellulose filters, and supernatant protein was precipitated by addition of trichloroacetic acid to a final concentration of 10% and incubation for 1 h at 0°C. Precipitates were collected by centrifugation for 20 min at 125,000 × g at 4°C, washed with ice-cold acetone, and air dried. For analysis of total bacterial protein, cells from overnight cultures were recovered by centrifugation for 5 min at 4,000 × g. Supernatant proteins and total bacteria were resuspended in sample buffer (12.5% glycerol, 4% sodium dodecyl sulfate [SDS], 2% β-mercaptoethanol, 0.01% bromophenol blue, 50 mM Tris-HCl [pH 6.8]), boiled for 5 min, and electrophoretically separated on SDS–12% polyacrylamide gels (21).
DNA biochemistry.
The fine mapping of transposon (Tn) insertions in ssaV was performed by DNA sequencing using primers corresponding to the I and O termini of mTn_5_ (16) as well as specific primers. Sequencing was performed by using dye terminator chemistry on a PE 377XL DNA sequencer.
For the generation of a nonpolar mutation in the ssaV gene, a 7.5-kb _Pst_I fragment from λ clone 7 (30) harboring ssaV was isolated and ligated into the _Pst_I site of pUC18. This construct was linearized at the _Eco_RV site in the ssaV gene and ligated with a 0.8-kb _Hin_cII fragment containing the aphT cassette (lacking transcriptional terminators) from plasmid pSB315 (12). The direction of transcription of the aphT gene within the disrupted ssaV gene was confirmed by a _Hin_dIII/Cla_I double digest and a plasmid selected which contains the aphT gene transcribed in the same direction as the ssaV gene. The disrupted ssaV gene was then isolated on an Eco_RI fragment and ligated into the suicide vector pGP704 (25) at the Eco_RI site to form plasmid pNP_ssaV. pNP_ssaV was transformed into S17-1 λ_pir and transferred by conjugation to S. typhimurium 12023 Nalr, using selection for kanamycin resistance. The correct integration of the ssaV::aphT mutation onto the chromosome was verified in individual exconjugants by restriction of genomic DNA with Cla_I and Hin_dIII in single digests and Southern hybridization. This mutation was transduced in the wild-type S. typhimurium 12023, and the resulting strain, NP_ssaV, was used for further studies. For the complementation of P9B7, ssaT was amplified by using primers SSAT-FOR (5′-CTAGGATCCGGCAGATAATGTTACG-3′), and SSAT-REV (5′-GCTGGATCCGCTCATACAGATGGAAAC-3′), and ssaTU was amplified by using primers SSAT-FOR and SSAU-REV (5′-GCTGGATCCATTTATGGTGTTTCGGTAG-3′), introducing Bam_HI restriction sites (underlined). Plasmids p_ssaT and p_ssaTU were generated by ligation of the respective _Bam_HI-digested PCR products to _Bam_HI-digested pACYC184.
Expression of recombinant SipC; biochemical and serological procedures.
For the expression of recombinant SipC, the sipC gene of S. typhimurium 12023 was amplified by using primers SIPC-FOR 5′-CTCGGATCCCGCCGCTTATTTA-3′ and SIPC-REV 5′-TGAAAGCTTAAGCGCGAATATTGCC-3′, introducing restriction sites for _Bam_HI and _Hin_dIII, respectively (underlined). A single PCR product of about 1.2 kb was obtained, digested with _Bam_HI and _Hin_dIII, and purified by electrophoresis on an agarose gel. The purified fragment was ligated into _Bam_HI- and _Hin_dIII-digested His tag fusion vector pQE30 (Qiagen, Hilden, Germany) to generate pJD10. E. coli M15[pREP] was transformed to ampicillin resistance by electroporation with pJD10. Expression of the His-tagged SipC fusion protein (recombinant SipC [rSipC]) from pJD10-harboring strains was detected with a Ni-alkaline phosphatase conjugate according to the instructions of the supplier (Qiagen). A highly expressing strain was selected and used for large-scale expression of rSipC. rSipC was purified by affinity chromatography on HiTrap chelating columns as instructed by the manufacturer (Pharmacia, Freiburg, Germany). The eluate was further purified by preparative SDS-polyacrylamide gel electrophoresis (PAGE), and a 42-kDa protein corresponding to rSipC was excised. rSipC was recovered from gel slices by electroelution.
For immunization, about 1 mg of rSipC was emulsified with complete and incomplete Freund’s adjuvants for primary and booster immunizations, respectively. A rabbit was immunized subcutaneously according to standard procedures (14), and antiserum was used without further modification. SipC was detected with the antiserum raised against rSipC after electrophoretic separation of proteins from culture supernatants or total cell fractions and transfer onto a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany), using a semidry blotting device (Bio-Rad). Bound antibody was visualized by using a secondary antibody-alkaline phosphatase conjugate according to standard procedures (14).
Reporter gene assays.
Expression levels of lacZ reporter gene fusions to SPI1 genes were assayed as described by Miller (24). Cultures of various S. typhimurium strains were grown overnight under conditions identical to those described for the analysis of secreted and cellular SipC. All reporter assays were performed at least in triplicate on different occasions.
Sensitivity assays.
Bacteria were grown to mid-logarithmic phase in LB broth, washed twice with RPMI 1640 (RPMI), and resuspended in RPMI at a concentration of approximately 108 CFU/ml. Gentamicin sensitivity assays were performed by adding an equal volume of RPMI containing twice the final concentration of gentamicin (0.62 μg/ml; Sigma) to bacterial suspensions and incubating the cells at 37°C for 1 h. To enumerate the number of viable bacteria present, an equal volume of RPMI without gentamicin was added to the bacterial suspensions, which were then incubated as described above. After incubation, the bacteria were harvested by centrifugation at 18,000 × g for 4 min, washed once with RPMI, and resuspended in RPMI. A dilution series was prepared in RPMI and plated onto LB or LB containing kanamycin (50 μg/ml) to enumerate CFU. Sensitivity to polymyxin B was assayed essentially as described by Roland et al. (29). Briefly, bacteria were grown to mid-logarithmic phase in LB broth, washed and resuspended in 0.5% tryptone saline at 104 CFU/ml, and incubated at 37°C for 1 h with or without polymyxin B (Sigma) at a final concentration of 500 ng/ml. After incubation, the test samples were diluted in saline and plated onto LB to determine the number of CFU, and the percent survival was calculated. Serum sensitivity assays were performed with pooled normal human serum as previously described (17).
RESULTS
Effects of mutations in SPI2 on a secreted effector protein of SPI1.
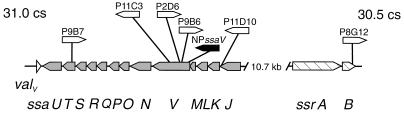
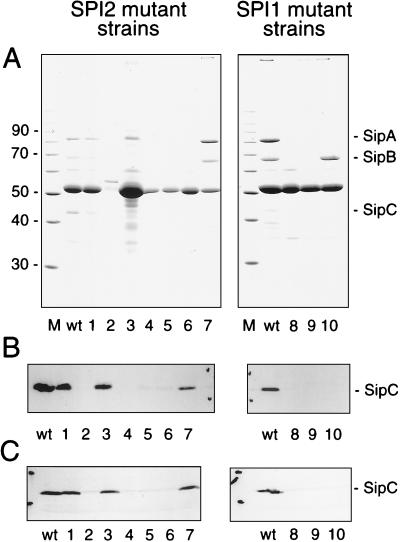
The positions and orientations of Tn insertions of mutant strains used in this study are indicated in Fig. 1. Analysis of proteins secreted into the growth medium by the S. typhimurium SPI2 mutant strains P9B7 (ssaT::mTn_5_) and P11C3 (ssaV::mTn_5_) revealed the absence or strong reduction in the amounts of the secreted SPI1 effector protein SipC (17). These SPI2 mutants are also reduced in the ability to invade cultured epithelial cells or cultured macrophages (17). To examine this phenomenon in greater detail, we expressed rSipC and raised antibodies against rSipC in rabbits. On Western blots, antiserum against rSipC reacted with a 42-kDa protein from precipitates of culture supernatants of S. typhimurium wild-type strain 12023. No reaction was observed with supernatants from cultures of EE638, a strain deficient in SipC (18) (data not shown) and a mutant (P7B12) from our STM collection harboring a mTn_5_ insertion in sipC (Fig. 2B). Furthermore, on Western blots SipC was not detected in culture supernatants of the SPI2 mutants P8G12 (ssrB::mTn_5_), P9B7 (ssaT::mTn_5_), P11C3 (ssaV::mTn_5_), and P11D10 (ssaJ::mTn_5_) and SPI1 mutants P4H2 (hilA::mTn_5_) and P6E11 (spaRS::mTn_5_). However, SipC was detected in culture supernatants of SPI2 mutants P2D6 (ssaV::mTn_5_), P9B6 (ssaV::mTn_5_), and NP_ssaV_ (ssaV::aphT). The detection by antiserum of SipC in culture supernatants of various strains was in accord with the presence or absence of SipC as detected by SDS-PAGE (Fig. 2A).
FIG. 1.
SPI2. The positions and orientations of insertions of mTn_5_ insertions (open arrows) and the aphT gene cassette (solid arrow) in mutant strains used in this study are shown. The transcriptional orientation of SPI2 genes encoding the type III secretion system apparatus (ssa) and the two-component regulatory system (ssr) is indicated by arrows. cs, centisomes.
FIG. 2.
Protein secreted by S. typhimurium and detection of SipC. (A) Secreted protein was precipitated from 2 ml of culture supernatant, separated by SDS-PAGE, and stained with Coomassie brilliant blue R-250. Positions of the SPI1 effector proteins SipABC (right) and the molecular weight marker (in thousands on the left) are indicated. (B) As for panel A except that protein was transferred onto nitrocellulose membranes after electrophoretic separation. The presence of SipC was detected with a polyclonal antiserum raised against rSipC. (C) Detection of SipC in total cell fractions of S. typhimurium. Equal amounts of protein (about 20 μg) of cells from overnight cultures were separated on SDS–12% polyacrylamide gels and subsequently transferred onto nitrocellulose membranes. SipC was detected after incubation with the rabbit antiserum against rSipC as described above. Lanes: M, marker; wt, wild-type S. typhimurium 1 to 7, SPI2 mutant strains P2D6, P11C3, P9B6, P11D10, P9B7, P8G12, and NP_ssaV_, respectively; 8 to 10, SPI1 mutant strains P4H2, P6E11, and P7B12, respectively.
Next we analyzed whether the absence of SipC in culture supernatants of SPI2 mutant strains was due to defective secretion of SipC via the type III secretion system or reduced synthesis of SipC in these strains. Antiserum against rSipC was used to detect SipC in pellets of cultures grown under inducing conditions for the expression of SPI1 genes (i.e., stationary phase, high osmolarity, and low oxygen) (3). Analysis of wild-type and strains carrying various mutations in SPI1 and SPI2 genes indicated highly reduced amounts of SipC in mutants harboring Tn insertions in SPI1 genes hilA (P4H2), spaRS (P6E11), and sipC (P7B12) and in the SPI2 mutant strains P8G12, P9B7, P11C3, and P11D10 (Fig. 2C). However, SipC was detected at levels comparable to those observed in pellets of wild-type cultures and SPI2 mutant strains P2D6, P9B6, and NP_ssaV_. The effect on SipC synthesis is not due to reduced growth rates or reduced protein synthesis levels in SPI2 mutants, since both parameters were comparable for the wild-type and SPI2 mutants (data not shown).
Effects of SPI2 mutations on the expression of SPI1 genes.
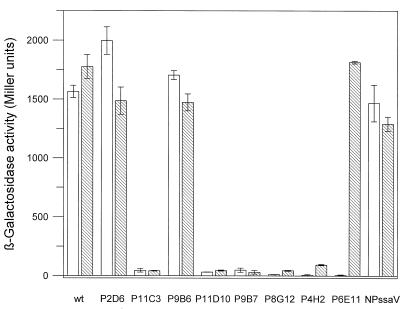
To assay the effect of SPI2 mutations on the expression of SPI1 genes, previously characterized fusions of lacZ to various SPI1 genes (2, 3) were transduced into various SPI2 and SPI1 mutants to generate a set of reporter fusion strains. The expression of the reporter β-galactosidase in cultures grown under conditions inducing for SPI1 expression (see above) was assayed. A Tn insertion in hilA (P4H2) reduced the expression of both prgK and sipC, while an insertion in spaRS (P6E11) affected the expression of only sipC (Fig. 3). Some mutant strains with Tn insertions in SPI2 genes encoding components of the type III secretion apparatus (P11C3, P11D10, and P9B7) or the two-component regulatory system (P8G12) showed reduced expression of reporter fusions to prgK and sipC (Fig. 3). The effects on the expression of both genes were similar. Other mutant strains with Tn insertions in ssaV (P2D6 and P9B6), as well as mutant NP_ssaV_ harboring a nonpolar insertions in ssaV, had levels of expression of prgK and sipC comparable to those of corresponding reporter fusions in a wild-type genetic background. Analysis of lacZ fusions to prgH and invF revealed an effect on expression similar to that shown for prgK and sipC (data not shown).
FIG. 3.
Effect of SPI2 mutations on expression of SPI1 genes. The expression of lacZ fusions to prgK (hatched bars) and sipC (open bars) was analyzed in various SPI1 and SPI2 backgrounds. Bacterial cultures were grown overnight under inducing conditions, and enzyme activity was determined as described in Materials and Methods. wt, wild type.
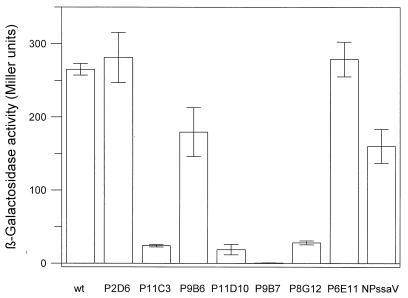
SPI2 mutations affect expression of the SPI1 regulator hilA.
Analysis of reporter fusions to sipC and prgK indicated that expression of genes in two different operons of SPI1 can be affected by SPI2 mutations, suggesting that these mutations affect other SPI1 genes involved in regulation of sipC and prgK. It has been demonstrated previously that the expression of SPI1 genes is under the control of the transcriptional activator HilA (2, 3). The expression of hilA was therefore analyzed in the presence of various SPI2 mutations. The SPI2 mutant strains P8G12, P9B7, P11C3, and P11D10 had largely diminished levels of hilA expression (Fig. 4). Again, very low levels of hilA expression were observed in mutants that had reduced levels of prgK and sipC expression. To analyze whether the effect of SPI2 mutations on SipC expression resulted from the reduced expression of hilA, we next performed complementation experiments in various mutant strains harboring pVV135 (constitutive expression of hilA) (3) or pVV214 (expression of hilA from the native promoter) (2). In accordance with a previous study (2), the hilA mutation of strain P4H2 was complemented by pVV214. However, SipC expression was not restored in mutant strain P11C3, P9B7, or P8G12 harboring either pV135 or pVV214 (data not shown).
FIG. 4.
Effect of SPI2 mutations on expression of hilA. The expression of a lacZ fusion to hilA was analyzed in various SPI1 and SPI2 backgrounds as indicated for Fig. 3. wt, wild type.
Effects of mutations in ssaV and ssaT.
Different mutations in ssaV produced different phenotypic effects. While the Tn insertion of mutant P11C3 resulted in a strong reduction of SPI1 gene expression, strains P2D6 and P9B6 with insertions at other positions of the same gene exhibited wild-type levels of SPI1 gene expression. To investigate the role of ssaV in more detail, a nonpolar mutation in ssaV was constructed. We analyzed the effect of the inactivation of ssaV on the virulence of S. typhimurium by determining the 50% lethal dose (LD50) in susceptible BALB/c mice. After intraperitoneal inoculation, an LD50 of 4.5 × 106 CFU was obtained for NP_ssaV_. This level of attenuation is comparable to SPI2 mutants with mTn_5_ insertions in other parts of the ssaK-U operon (30). However, synthesis of SipC and expression of SPI1 genes were not significantly affected in NP_ssaV_, showing that ssaV is not required for SPI1 secretion system function. We also observed that a mTn_5_ insertion in ssaT (strain P9B7) (Fig. 1) resulted in reduction of SPI1 gene expression. Since ssaT is the penultimate gene of the ssaK-U operon, this observation suggests that ssaT and/or ssaU are important for SPI1 gene expression. SsaT and SsaU are homologs of YscT and YscU of the virulence plasmid-encoded type III secretion system of Yersinia spp. (17). Both proteins contain several putative transmembrane helices and may form structural components of the type III secretion system (1, 5). To further analyze the effect of SsaT and/or SsaU on expression of SPI1 genes, complementation experiments were performed with plasmids p_ssaT_ and p_ssaTU_, which constitutively express ssaT and ssaTU, respectively, from the Tetr promoter of pACYC184. The virulence of strain P9B7[p_ssaTU_] in BALB/c mice after intraperitoneal infection with 104 CFU was restored, indicating that the function of SPI2 was complemented by p_ssaTU_. The presence of p_ssaT_ or p_ssaTU_ had no effect on SipC expression in the wild-type strain; however, SipC expression was not restored in mutant strains P9B7 and P11C3 harboring either p_ssaT_ or p_ssaTU_ (data not shown).
Sensitivity of SPI2 mutants to serum, gentamicin, and polymyxin B.
In a previous study (17), we found that some mutations in the ssaV, ssaT, and ssaJ genes resulted in altered resistance to killing by complement, whereas other resulted in wild-type levels of resistance. These data suggested that mutations in these genes caused a perturbation of the cell wall. Accordingly, we investigated whether mutations in these and other SPI2 genes could affect sensitivity to serum, gentamicin, and polymyxin B, since these compounds affect or need to cross the cell wall for activity. Mutations in the ssaT and ssr genes cause increased sensitivity to killing by complement and gentamicin, while some mutations in ssaV and ssaJ show enhanced resistance to killing by complement and wild-type levels or enhanced resistance to gentamicin. All of the SPI2 mutants analyzed were more sensitive to polymyxin B than the wild-type strain, with ssaT mutant P9B7 being the most sensitive to polymyxin B (Table 2).
TABLE 2.
Sensitivity of selected SPI2 mutants to killing by complement, gentamicin, and polymyxin B
| Strain | Mutation | Sensitivity (mean log10 kill ± SD) | Polymyxin B sensitivity (% survival) | ||
|---|---|---|---|---|---|
| Seruma | Gentamicinb | ||||
| Reference 17 | This study | ||||
| 12023 | Wild type | 0.356 ± 0.15 | 0.176 ± 0.133 | 2.115 ± 0.08 | 62.8 ± 13.2 |
| P3F4 | ssrA::mTn_5_ | 1.649 ± 0.48 | 5.125 ± 0.15 | 12.4 ± 3.9 | |
| P8G12 | ssrB::mTn_5_ | 2.909 ± 0.04 | 4.416 ± 0.15 | 25.4 ± 13.3 | |
| P2D6 | ssaV::mTn_5_ | 0.034 ± 0.01 | 2.076 ± 0.06 | 13.1 ± 6.1 | |
| P11C3 | ssaV::mTn_5_ | 0.045 ± 0.02 | 1.743 ± 0.01 | 17.8 ± 6.8 | |
| NP_ssaV_ | ssaV::aphT | 0.184 ± 0.03 | 2.244 ± 0.14 | 50.1 ± 3.2 | |
| P11D10 | ssaJ::mTn_5_ | 0.085 ± 0.05 | 1.946 ± 0.07 | 43.2 ± 6.9 | |
| P9B7 | ssaT::mTn_5_ | 1.652 ± 0.34 | 4.257 ± 0.09 | 1.8 ± 1.7 |
DISCUSSION
We previously showed that certain mutations in SPI2 affect the ability of S. typhimurium to invade cultured epithelial cells and cultured macrophage-like cells, and it has been suggested that this is a secondary effect due to a decreased secretion of SPI1 effector proteins (17). Here we have confirmed and extended these findings by showing that the effect on SPI1 functions is correlated to reduced expression of SPI1 genes in certain SPI2 mutant strains. Collazo and Galan (7) have demonstrated that a functional type III secretion system of SPI1 is required for the secretion of SPI1 effector proteins. According to the model proposed by Bajaj et al. (2), HilA is the key local regulatory protein for the expression of SPI1 genes, and it has been demonstrated that the expression of SPI1 operons encoding type III secretion system components (such as PrgK) and secreted effector proteins (such as SipC) is under the control of HilA (2, 3). SirA and PhoPQ, encoded by genes outside the SPI1 and SPI2 loci, also regulate the expression of hilA (19). A possible explanation for the reduced expression of sipC may be the reduced expression of hilA resulting from mutations in SPI2. However, our results indicate that this is unlikely since regulated or constitutive expression of hilA in trans did not complement the effects of mutations in SPI2 on sipC expression. If the expression of SPI1 genes is not directly dependent on the amount of HilA present in the cell, SPI2 mutations may affect other proteins also regulating SPI1 gene expression. Alternatively, mutations in SPI2 genes may affect the activity of promoters of SPI1 genes more directly. It has been demonstrated that the activity of HilA is modulated by environmental factors (3), but the molecular nature of this modulation has not been elucidated. The recognition of such environmental signals affecting HilA activity may be disturbed in strains carrying certain SPI2 mutations. This hypothesis is supported by the observation that certain mutations in SPI2 have pleiotropic effects resulting in increased sensitivity to complement, gentamicin, and polymyxin. As all three of these antimicrobial agents require an interaction with the bacterial cell surface or membrane components to be effective (for review, see reference 20), this finding suggests that the cell membrane or surface is perturbed in these mutants. This may be due to mutant SPI2 components present in or spanning the cell membrane or to the lack of SPI2 components. However, it should be noted that not all SPI2 mutants with reduced expression of SPI1 genes show increased sensitivity to antimicrobial agents. The global regulation by DNA supercoiling is another regulatory factor for SPI1 gene expression (11). If such regulation by DNA topology is disturbed in the background of SPI2 mutations, this effect could act in parallel on several promoters within SPI1. Under such conditions, SPI1 promoters may not be active even in the presence of HilA. Alternatively, some products of SPI2 genes may directly affect the activity of HilA. The expression of SPI2 genes is largely reduced in an ssrB mutant strain (P8G12) (14a, 31). This mutation also led to reduced expression of SPI1 genes, indicating that the presence of one or more SPI2 proteins is required for expression of SPI1 genes.
The effect of a mutation in ssaT on S. typhimurium virulence was complemented by constitutive expression of ssaTU in trans. Presence of p_ssaTU_ in the wild-type background did not influence SipC expression; however, the effect of the ssaT mutation in strain P9B7 on SPI1 gene expression was not complemented by p_ssaTU_. This result indicates that the mutation in ssaT may have a dominant effect on the expression of SPI1 genes that cannot be complemented by the addition of the wild-type allele. Alternatively, the levels of SsaTU required for the complementation of the SPI2 virulence function may be different from the amounts of SsaTU required for SPI1 function. There is no obvious explanation for the observation that only certain SPI2 mutations in the ssaK-U operon affect expression of SPI1 genes. It has been observed that the transcriptional terminator of the aph gene of Tn_5_ can cause polar effects (13). The Tn insertion may also introduce promoters activating the transcription of genes downstream of the Tn insertion site (4). Either effect of a Tn insertion could result in a disturbance of the ratio between SPI2 subunits encoded by the ssaK-U operon. Further work is needed to determine if the lack of components of the SPI2 secretion system or the presence of components of the SPI2 secretion system in unbalanced amounts affects the expression of SPI1 genes.
The similar intraperitoneal LD50s obtained for the ssrA::mTn_5_ strain P3F4, ssaT::mTn_5_ strain P9B7 (30), and mutant NP_ssaV_ (this study) demonstrate that the effect of antimicrobial agents such as complement, gentamicin, and polymyxin are very likely to be a secondary consequence of mutations in SPI2 genes, and their physiological relevance is uncertain. However, even if these effects are secondary, they have important implications for the study of SPI2 mutants, particularly in cell culture invasion or replication assays that use gentamicin over long periods of time to kill extracellular bacteria.
In conclusion, our observation of the interaction between virulence loci in S. typhimurium suggests that an intact SPI2 secretion system may be required for normal expression of genes in SPI1. Present understanding of the regulation of gene expression of SPI1 and SPI2 is not sufficient to explain the molecular basis of effects of SPI2 mutations on SPI1 function. However, the potential regulatory interaction described here should be considered with respect to experiments designed to understand the function of the type III secretion system of SPI1 and the role of SPI1 in pathogenesis.
ACKNOWLEDGMENTS
This project was supported by the Deutsche Forschungsgemeinschaft (grant He 1964/2-2 to M.H.) and by a grant from the Medical Research Council (United Kingdom) to D.W.H.
We are indebted to C. A. Lee (Harvard University) for providing strains and plasmids and to J. E. Galan (Yale University) for plasmid pSB315. We especially thank J. Heesemann (Munich, Germany) for support and discussion.
REFERENCES
- 1.Allaoui A, Woestyn S, Sluiters C, Cornelis G R. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 4.Berg D E, Weiss A, Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980;142:439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergman T, Erickson K, Galyov E, Persson C, Wolf-Watz H. The lcrB (yscN/U) gene cluster of Yersinia pseudotuberculosis is involved in Yop secretion and shows high homology to the spa gene clusters of Shigella flexneri and Salmonella typhimurium. J Bacteriol. 1994;176:2619–2626. doi: 10.1128/jb.176.9.2619-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang A, C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collazo C M, Galan J E. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 9.Galan J E. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 10.Galan J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan J E, Curtiss R., III Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun. 1990;58:1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galan J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harayama S, Bollinger J, Iino T, Hazelbauer S L. Characterization of the mgl operon of Escherichia coli by transposon mutagenesis and molecular cloning. J Bacteriol. 1983;153:408–415. doi: 10.1128/jb.153.1.408-415.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 14a.Hensel, M., T. Nikolaus, and J. Deiwick. Unpublished data.
- 15.Hensel M, Shea J E, Bäumler A J, Gleeson C, Blattner F R, Holden D W. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 17.Hensel M, Shea J E, Raupach B, Monack D, Falkow S, Gleeson C, Holden D W. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 18.Hueck C J, Hantman M J, Bajaj V, Johnston C, Lee C A, Miller S I. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 19.Johnston C, Pegues D A, Hueck C J, Lee C A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 20.Joiner K A. Complement evasion by bacteria and parasites. Annu Rev Microbiol. 1988;42:201–230. doi: 10.1146/annurev.mi.42.100188.001221. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maloy S R, Steward V L, Taylor R K. Genetic analysis of pathogenic bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 24.Miller J H. A short course in bacteria genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 25.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills D M, Bajaj V, Lee C A. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 27.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Raupach, B. Unpublished data.
- 29.Roland K L, Martin L E, Esther C R, Spitznagel J K. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]