Activator-Specific Requirement of Yeast Mediator Proteins for RNA Polymerase II Transcriptional Activation (original) (raw)
Abstract
The multisubunit Mediator complex of Saccharomyces cerevisiae is required for most RNA polymerase II (Pol II) transcription. The Mediator complex is composed of two subcomplexes, the Rgr1 and Srb4 subcomplexes, which appear to function in the reception of activator signals and the subsequent modulation of Pol II activity, respectively. In order to determine the precise composition of the Mediator complex and to explore the specific role of each Mediator protein, our goal was to identify all of the Mediator components. To this end, we cloned three previously unidentified Mediator subunits, Med9/Cse2, Med10/Nut2, and Med11, and isolated mutant forms of each of them to analyze their transcriptional defects. Differential display and Northern analyses of mRNAs from wild-type and Mediator mutant cells demonstrated an activator-specific requirement for each Mediator subunit. Med9/Cse2 and Med10/Nut2 were required, respectively, for Bas1/Bas2- and Gcn4-mediated transcription of amino acid biosynthetic genes. Gal11 was required for Gal4- and Rap1-mediated transcriptional activation. Med11 was also required specifically for MFα1 transcription. On the other hand, Med6 was required for all of these transcriptional activation processes. These results suggest that distinct Mediator proteins in the Rgr1 subcomplex are required for activator-specific transcriptional activation and that the activation signals mediated by these Mediator proteins converge on Med6 (or the Srb4 subcomplex) to modulate Pol II activity.
Regulation of mRNA synthesis by transcriptional activator proteins requires many diverse regulatory proteins collectively called transcriptional coactivators (for reviews, see references 2, 17, and 40). The TATA binding protein-associated factors (TAFIIs), which compose the TFIID complex, and the multisubunit Mediator complex are the two major coactivators that enable the basal transcription machinery to respond to gene-specific transcriptional regulatory proteins.
TAFIIs were initially identified in human and Drosophila as essential factors for transcriptional activation in a reconstituted transcription system (12, 33). Biochemical analysis of TFIID revealed a modular structure in which a large TAFII subunit, acting as a scaffold, binds to several distinct TAFII subunits, each of which interacts with specific transcriptional activator proteins (4). However, depletion or inactivation of TAFIIs from the yeast Saccharomyces cerevisiae caused no obvious defect in transcriptional activation in vivo (28, 41). Therefore, it was proposed that TAFIIs function as essential cofactors for transcription of only a subset of genes, rather than as general targets of transcriptional activators (1, 35).
In contrast to the limited requirement for TAFIIs, a second coactivator complex, the Mediator complex, appears to be required for the transcription of most RNA polymerase II (Pol II)-transcribed genes. The Mediator complex is required not only for transcriptional activation but also for the stimulation of basal transcription and higher carboxy-terminal domain (CTD) phosphorylation efficiency by TFIIH (18). Mediator is tightly associated with the CTD of Pol II and is composed of the Med proteins (24, 29); Gal11, Rgr1, Sin4, Hrs1, and Rox3 (9, 18, 26, 39); and the Srb family of proteins. Mediator components with genetically similar phenotypes are physically associated, thus forming two major Mediator subcomplexes, the Srb4 subcomplex and the Rgr1 subcomplex (23). The Srb4 subcomplex contains all of the genetically dominant Srb proteins (Srb2, -4, -5, and -6) and Med6 and appears to modulate Pol II activity through its interaction with the CTD. The Srb4 subcomplex was successfully reconstituted in vitro with recombinant Med6 and Srb proteins (19), and the functional interactions between components of this subcomplex were shown genetically by the suppressor relationships among the SRB4, SRB6, and MED6 genes (22, 23).
The remaining Mediator components form the Rgr1 subcomplex, which plays an apparent role in activator-specific functions. At least one activator-specific module, the Gal11 module, which contains Gal11, Sin4, and Hrs1, was shown to interact physically with the C-terminal domain of the Rgr1 protein. Mutations in each of the components of the Gal11 module were shown to yield similar mutant phenotypes and to affect transcriptional regulation of the same subset of genes (15, 26, 32, 37). These results suggest that the Gal11 module functions in the receiving end of signals from a subset of gene-specific transcriptional regulators. Other members of the Rgr1 subcomplex interact with Rgr1 through regions other than its C-terminal domain (23). However, whether these polypeptides form a module(s) with a specific regulatory function as do the Gal11 module components remains to be examined.
In order to elucidate the mechanism by which Mediator functions to bridge gene-specific activators and Pol II, it is important to address how activator specificity is achieved and to decipher which Mediator proteins are required for specific transcriptional activation events. Despite the fact that a number of new Mediator genes have been reported recently (29), the precise composition of the Mediator complex remains elusive. Therefore, we purified the Mediator complex to homogeneity from S. cerevisiae with the use of an anti-Rgr1 antibody column and thus were able to identify all of the remaining Mediator proteins (Med9/Cse2, Med10/Nut2, and Med11) that had escaped earlier identification efforts. Here we report the functional analysis of these new Mediator genes and present evidence that defines the activator-specific requirements of individual Mediator proteins tethered to Rgr1. Our results reveal the specific functions of each Mediator component in the relay of gene-specific activator signals to Pol II.
MATERIALS AND METHODS
Protein purification.
A whole-cell extract from a strain containing His-tagged Med6 was fractionated according to the procedure described by Kim et al. (18) with the following modifications. We employed an immunoaffinity purification step with the use of an antibody to Rgr1 in order to remove all of the contaminating polypeptides that copurified with holopolymerase through the preceding chromatographic steps. Anti-Rgr1 antiserum (1 ml) was conjugated with protein G-agarose beads (1 ml) (GIBCO BRL, Gaithersburg, Md.), and the resulting affinity resin was used to purify holopolymerase from 2 ml of the MonoQ fraction (protein concentration, 0.6 mg/ml) according to the procedure described by Lee and Kim (23). After separation of the affinity column-eluted proteins on a sodium dodecyl sulfate (SDS)-polyacrylamide (13%) gel, proteins were stained with Coomassie blue, and previously unidentified low-molecular-size Mediator proteins (15, 19, and 20 kDa) were excised for peptide sequencing.
Peptide sequencing by ion trap MS.
The excised protein bands were subjected to in-gel reduction, _S_-carboxyamidomethylation, and tryptic digestion (Promega, Madison, Wis.), and a 10% aliquot of the resultant mixture was analyzed as follows. Sequence information was determined by capillary (180-μm by 15-cm column; LC Packings, Amsterdam, The Netherlands) reverse-phase chromatography coupled to the electrospray ionization source of a quadrupole ion trap mass spectrometer (Finnigan LCQ, San Jose, Calif.) (30). Peptides were eluted with a gradient of 10.8 to 41.6% acetonitrile in 0.1% acetic acid–0.02% trifluoroacetic acid. The instrument was programmed to acquire successive sets of three scan modes consisting of full-scan mass spectrometry (MS) over the m/z range of 395 to 1,118 μ, followed by two data-dependent scans on the most abundant ion in that full scan. These data-dependent scans allowed the automatic acquisition of a high-resolution (zoom scan) spectra to determine charge state and exact mass and of MS-MS spectra for the peptide sequence information. Intepretation of the resulting MS-MS spectra of the peptides was facilitated by searching the National Center for Biotechnology Information nr and dbest databases with the algorithm Sequest (7), followed by manual inspection.
Cloning and disruption of Mediator genes.
The flanking regions of MED9 (bp −239 to +3 and +277 to +559), MED10 (−438 to −54 and +203 to +623), and MED11 (−473 to −138 and +519 to +851) (the translation initiation site is +1) were amplified from yeast genomic DNA by PCR. Each pair of amplified 5′ upstream and 3′ downstream regions was cloned into the _Bam_HI and _Hin_dIII sites of pRS316 to construct the gap repair plasmids pSJ1 (MED9), pSJ2 (MED10), and pSJ3 (MED11). In order to construct gene disruption plasmids, TRP1 was introduced into the _Eco_RI site between the cloned 5′ and 3′ regions of the Mediator genes on these gap repair plasmids to yield pSJ1-1, pSJ2-1, and pSJ3-1. Wild-type alleles of the Mediator genes were cloned with the use of the pSJ1, -2, and -3 gap repair plasmids as described by Lee et al. (24). Sequencing analysis of the recovered inserts confirmed the isolation of wild-type alleles of MED9, MED10, and MED11 on pSJ4, -5, and -6, respectively. Mediator gene deletion strains were made according to the procedure described by Lee et al. (24) with the use of the gene disruption plasmids to yield strains YSJ9M, YSJ10S, and YSJ11S (Table 1).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| YPH500 | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 |
| YPHBAS | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1, p2100a (HIS3), pSH18-34b (URA3) |
| YPHG4 | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1, pHYC1 (66)c (URA3) |
| YSJ9M | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed9::TRP1 |
| YSJ9R | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed9::TRP1/MED9 on pRS316 (URA3) |
| YSJ9BAS | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed9::TRP1, p2100 (HIS3), pSH18-34 (URA3) |
| YSJ9G4 | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed9::TRP1, pHYC1 (66) (URA3) |
| YSJ10S | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed10::TRP1/MED10 on pRR316 (URA3) |
| YSJ10M | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed10::TRP1/ts mutant med10 on pRS313 (HIS3) |
| YSJ10R | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed10::TRP1/ts mutant med10 on pRS313 (HIS3)/MED10 on pRS316 (URA3) |
| YSJ10HA | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed10::TRP1/HA-MED10 on pRR315 (LEU2) |
| YSJ10BAS | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed10::TRP1/ts mutant med10 on pRS315 (LUE2), p2100 (HIS3), pSH18-34 (URA3) |
| YSJ10G4 | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed10::TRP1/ts mutant med10 on pRS315 (LUE2), pHYC1 (66) (URA3) |
| YSJ11S | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed11::TRP1/MED11 on pRR316 (URA3) |
| YSJ11M | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed11::TRP1/med11 ts mutation on pRS313 (HIS3) |
| YSJ11R | MATα ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 Δmed11::TRP1/med11 ts mutation on pRS313 (HIS3)/MED11 on pRS316 (URA3) |
Isolation of ts mutants.
The temperature-sensitive (ts) mutants for MED10 and MED11 were generated according to the procedure described by Lee et al. (24) with the following modifications. The MED10 and MED11 mutant PCR products were cotransformed into YSJ10S and YSJ11S, respectively, with a linearized, _HIS3_-based pSJ7 or pSJ8 plasmid containing 250 bp of flanking sequences from the corresponding MED genes at their termini. Transformants were selected against 5-fluoro-orotic acid to remove the corresponding wild-type MED gene. The 5-fluoro-orotic acid-resistant colonies were grown on yeast extract-peptone-dextrose (YPD) medium, and mutants showing ts lethality were isolated.
Antibody preparation and immunoprecipitation experiments.
MED9 and MED11 open reading frames (ORFs) were inserted into the _Bam_HI and _Xho_I sites of the pGEX-4T-1 vector (Pharmacia, Uppsala, Sweden) to construct the glutathione _S_-transferase (GST)–Med9 and glutathione _S_-transferase–Med11 fusion protein expression constructs, respectively. Polyclonal antibodies to the fusion proteins were raised in rats and affinity purified as described by Lee et al. (24). Instead of raising antibodies to Med10, we introduced a double hemagglutinin (HA) tag at the N-terminal end of Med10 by PCR with primers that contained two copies of the HA epitope sequence (p10HA-2 [5′-ATC CAT ATG ATG TTC CAG ATT ATG C-3′] and p10HA-3 [5′-GTC AGG TAC GTC GTA AGG GTA AGC ATA ATC TGG-3′]) and cloned the resulting PCR product into the _Bam_HI and _Hin_dIII sites of pRS315 to make pSJ9. The wild-type copy of the MED10 gene on pRS316 in strain YSJ10S was replaced with pSJ9 to make a strain (YSJ10HA) that contained only the HA-tagged version of Med10. Immunoprecipitation experiments were performed as described by Lee and Kim (23).
Differential display of Mediator mutant mRNAs.
Wild-type and mutant yeast cells were grown on YP-glucose medium to early exponential phase at 30°C (_A_600 = 0.3 to 0.4) and were then allowed to grow for an additional 2.5 h at 37°C. The cells were then harvested, and poly(A)+ RNA was prepared. Both the wild-type and mutant poly(A)+ RNAs were converted into cDNAs with nine two-base-anchored oligo(dT) primers (T1 to T9) to subdivide the poly(A)+ RNA population. The subdivided cDNA populations were amplified by PCR in the presence of 10-nucleotide arbitrary primers (AP1 to AP10) by using the Delta RNA Fingerprinting Kit (Clontech, Palo Alto, Calif.). From PCR experiments with 90 different primer combinations, an average of 6,300 amplified PCR bands were visualized for each mRNA preparation. Differentially displayed PCR bands were eluted and amplified by a second round of PCR and then used as probes in groups of three for Northern analysis. For PCR bands that yielded hybridization signals with different intensities for wild-type and mutant mRNAs, we cloned the PCR fragments into a pGEM-T Easy vector (Promega). We then repeated the Northern blot analysis with individual clones as the probes for genes differentially expressed in strains carrying the Mediator mutations. For those clones identified as positive by this procedure, we determined the nucleotide sequences.
RNA preparation and analysis.
Cells were grown in YPD medium to early exponential phase at 30°C, collected by centrifugation, washed with water, and resuspended with an equal volume of prewarmed (37°C) synthetic complex medium lacking lysine (for amino acid starvation) or YP-galactose medium (for galactose induction). After the medium shift, cells were allowed to grow for additional 2.5 h at 37°C, after which mRNA was prepared as described previously (24). In order to prepare the probes for Northern analysis, DNA fragments for the HIS4 gene (from bp +605 to +1804) and the MFα1 gene (from bp +3 to +498) were generated by PCR amplification with appropriate primers, and their sequences were verified by nucleic acid sequencing. For generation of a GAL1 probe, an oligonucleotide complementary to the GAL1 coding region (24) was synthesized and end labeled with T4 polynucleotide kinase and [γ-32P]ATP. Specifically hybridized signals were quantitated with the use of a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and the associated software. When necessary, the filter was stripped by incubation in a 0.1% SDS solution for 10 min at 90°C and rehybridized with different probes.
RESULTS
Cloning of MED9, MED10, and MED11.
The enormous size of the Mediator complex and its intrinsic binding affinity for many transcription factors have made it difficult to define its precise composition. This ambiguity hindered identification of the Mediator proteins required specifically for the transcriptional activation of distinct groups of genes. Therefore, we purified the Mediator-Pol II complex (holopolymerase) to homogeneity and determined the identities of all of the Mediator subunits.
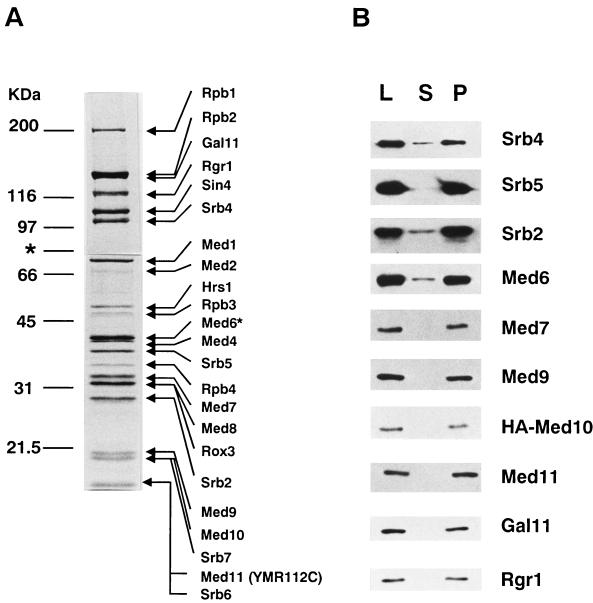
We supplemented our earlier fractionation scheme (18) with an anti-Rgr1 antibody column chromatography to obtain a highly purified holopolymerase fraction (see Materials and Methods). The holopolymerase preparation that was eluted from the anti-Rgr1 antibody column contained no additional known transcription factors (for example, TAFs, the Swi/Snf proteins, and general transcription factors were absent from the preparation) (Fig. 1A and data not shown). However, we found a previously unidentified protein band of 20 kDa, which we named Med9. In addition, we noticed that the Coomassie blue staining intensities of the 19- and 15-kDa protein bands (which correspond to Srb7 and Srb6, respectively) were twice what one would predict for the stoichiometric amount (data not shown). We named these previously unidentified proteins of 19 and 15 kDa Med10 and Med11, respectively. The peptide sequences that we obtained by ion trap mass spectrometry from the 15-, 19-, and 20-kDa protein bands originated from a total of five proteins, including Srb6 and Srb7, suggesting that no additional Mediator proteins were present.
FIG. 1.
Polypeptide composition of the Mediator-Pol II complex. (A) SDS-polyacrylamide gel electrophoresis of the holopolymerase complex immunopurified on an anti-Rgr1 antibody column. Molecular size marker proteins are indicated on the left, and the Pol II and Mediator components, including the new Mediator proteins (Med9, Med10, and Med11), are indicated on the right. For better resolution of its components, holopolymerase was separated on SDS–7.5% and –13% polyacrylamide gels, and the boundary of the two protein gels is marked with an asterisk. Med6*, histidine-tagged Med6. (B) Immunoblot analysis of the coimmunoprecipitation of the Med proteins. The HA-Med10 holopolymerase fraction was immunoprecipitated with an anti-HA monoclonal antibody (12CA5). Equivalent amounts of load (L), supernatant (S), and pellet (P) from the immunoprecipitation were blotted and probed with the antibodies indicated at the right.
A database search revealed that Med9 is identical to Cse2. A cse2 mutation (cse2-1) was originally isolated in a genetic screen for mutations that affect chromosome segregation (42). However, the role that CSE2 plays in chromosomal segregation is not known. We also learned that Med10 is identical to Nut2 (38). A nut2 mutation bypasses the Swi4 requirement only in the context of a lacZ reporter fused to a minimal HO promoter. Unlike Med9 and Med10, Med11 is a novel protein (ORF YMR112C) without sequence similarity to any known proteins.
The association of these polypeptides with holopolymerase was confirmed by their cofractionation throughout the holopolymerase purification procedure (data not shown). No other purification fractions contained traceable amounts of the Med proteins. In order to examine whether Med9, Med10, and Med11 are all present in a single complex with other Mediator proteins, Med10 was tagged with an HA epitope, and the holopolymerase fraction (MonoQ) from the HA-Med10 yeast strain (YSJ10HA) was immunoprecipitated with an anti-HA monoclonal antibody (12CA5). Immunoblot and silver staining analyses showed that Med9, Med11, and other holopolymerase components were immunoprecipitated together with HA-Med10 (Fig. 1B and data not shown). Immunoprecipitation with anti-Med9 antibody yielded identical results (data not shown). Therefore, Med9/Cse2, Med10/Nut2, and Med11 are genuine components of the Mediator complex.
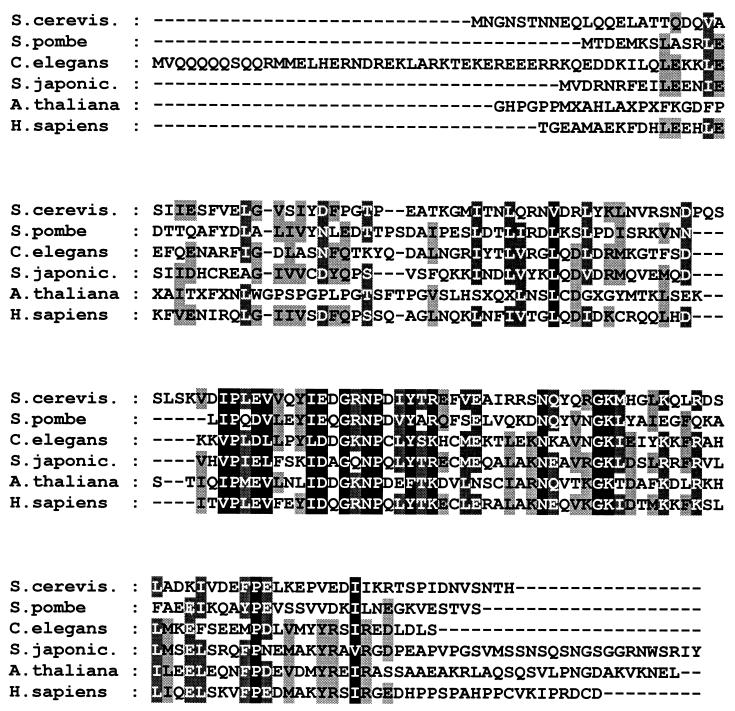
A search for homologs of these newly identified Mediator proteins found ORFs similar to that for yeast Med10 in human, Arabidosis, Caenorhabditis elegans, Schistosoma japonicum, and Schizosaccharomyces pombe databases. Comparison of yeast Med10 with other Med10 homologs revealed 50 to 35% sequence similarity (24 to 17% identity) (Fig. 2). Recently, human Med10 was identified independently as a component of a Mediator component (Srb7, Med6, and Rgr1)-containing human complex that exhibits both coactivator and repressor functions in vitro (34, 36). Whether this human complex is functionally equivalent to the yeast Mediator is not proven yet, but the identification of multiple human homologs in a complex suggests that at least some of the regulatory function of Mediator is conserved throughout evolution.
FIG. 2.
Sequence alignment of Med10 homologs. Protein sequences of Med10 homologs from S. cerevisiae (GenBank accession no. U25840), S. pombe (2226420), C. elegans (1176601), S. japonicum (AA661070), Arabidopsis thaliana (AA042215), and human (AA429956) are aligned. Identical amino acids (black boxes), similar amino acids (shaded boxes), and gaps in the sequence (dashed line) are indicated.
Isolation of med9, med10, and med11 mutants.
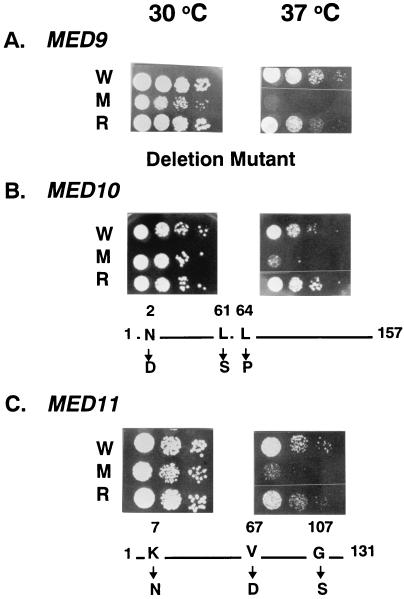
To conduct further studies on the newly discovered Mediator proteins, we generated mutant forms of each of the encoding genes. Because med9/cse2 was dispensable for cell viability (42), we deleted the whole MED9/CSE2 coding region from a haploid cell to make a med9/cse2 null strain (YSJ9M). As no such information was available for MED10 and MED11, we first examined whether they were essential for cell viability. We generated MED10 and MED11 deletions in individual diploid yeast strains. Tetrad analysis of both deletion strains gave only two viable spores that did not bear the TRP1 deletion marker, demonstrating that both genes are essential for cell viability (data not shown). Therefore, we isolated ts med10-1 and med11-1 mutants as described in Materials and Methods. The ts phenotypes were caused by specific mutations in the corresponding Mediator genes such that introduction of a wild-type copy of the gene rescued the growth defect at the restrictive temperature (Fig. 3). Mutation site analysis revealed three amino acid changes for both the med10-1 (N2D, L61S, and L64P) and med11-1 (K7N, V67D, and G107S) alleles.
FIG. 3.
ts phenotypes of the newly identified Mediator mutants. Yeast strains were spotted in duplicate on YPD agar plates, and each plate was incubated for 3 days at either the permissive (30°C) or nonpermissive (37°C) temperature. The temperature sensitivities of the wild-type strains (W), Mediator mutant strains (M), and mutant strains transformed with the corresponding wild-type Mediator gene (rescued [R]) were compared for med9 null (A), med10 ts (B), and med11 ts (C) mutants. At the bottom of each panel, the type of mutations and the mutated amino acids and their positions are shown.
Identification of genes affected by each Mediator mutation.
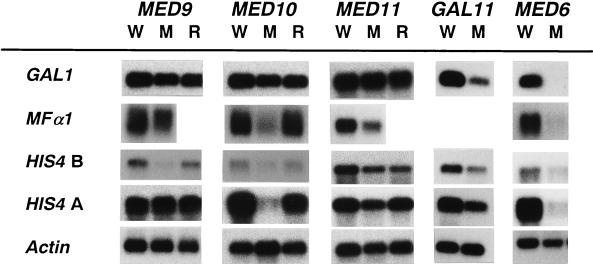
In order to identify the gene-specific functions of the newly identified Mediator proteins, we searched for genes whose expression was affected specifically by individual Mediator mutations. Differential display analyses of mRNA preparations from wild-type and Mediator mutant strains (see Materials and Methods) revealed that 37, 19, and 31 RNA fragments were preferentially amplified from wild-type mRNA compared with amplified med9, med10, and med11 mutant mRNAs, respectively. Northern blot analysis with the differentially displayed fragments as probes revealed a total of five transcripts (two for med9, three for med10, but none for med11) that were reduced at least fivefold in the mutant strains (Fig. 4). We also found 50 RNA fragments that were amplified only from the mutant mRNA preparations, but none of them showed more than a twofold difference in expression between wild-type and mutant strains (data not shown).
FIG. 4.
Northern analysis of genes isolated from differential display of Mediator mutant mRNAs. Wild-type (W) and mutant (M) mRNAs for each of the Mediator genes indicated at the top were prepared from yeast cells grown in YPD at the nonpermissive temperature. Each mRNA blot was hybridized with the probe indicated at the left. As an RNA loading control, actin transcript levels are shown.
Two genes, ARG4 and YGR260W, were identified as targets of Med9 regulation. YGR260W encodes a protein that is homologous to allantotate permease (DAL5). The ARG4 and YGR260W transcripts were reduced 8- and 10-fold in the med9 mutant, respectively (Fig. 4), and these transcriptional defects were rescued by introducing a wild-type copy of MED9 (data not shown). We also examined whether the med9 mutant strain was defective for transcription of genes involved in chromosomal segregation; however, transcription of a number of functionally related genes, including MIF2 (3), YMR092C and YNR047W (14), YLR457C and CBF1 (27), CHL4 (20), and CIN1 (13), was not compromised by the med9 mutation (data not shown). Expression of SCM2, the tryptophan permease gene, which was identified as a high-copy suppressor of the cold sensitivity phenotype of cse2-1 (5), also was not altered in the med9 mutant. These results indicate that Med9/Cse2 regulates the expression of genes involved in amino acid biosynthesis and that the med9 mutant phenotype may result from compromised amino acid synthesis.
Differential display experiments for the med10 mutant mRNA preparation also identified three differentially expressed genes, two amino acid biosynthetic genes (HIS4 and LYS20) and a novel gene (YGL117W). HIS4, LYS20, and YGL117W transcripts were specifically reduced 5- to 10-fold by the defective med10 activity (Fig. 4). Because both the med9 and med10 mutants showed defects in the transcription of genes involved in amino acid biosynthesis, we examined whether MED9 and MED10 have common transcriptional regulation targets. Indeed, the levels of ARG4 and YGR260W transcripts were reduced 5- and 10-fold in the med10 mutant, respectively (Fig. 4). Similarly, the med9 mutant was also defective (a three- to eightfold reduction) in the transcription of the genes affected by the med10 mutation (Fig. 4). These results revealed that both Med9/Cse2 and Med10 are required for the regulation of certain amino acid biosynthetic genes.
Gene-specific transcriptional regulatory functions of individual Mediator genes.
Despite the well-documented requirement for Mediator genes (for example, MED6, GAL11, SIN4, HRS1, and RGR1) in the transcriptional activation of genes involved in carbon source metabolism and mating type specification (2, 21, 32), the genes identified as regulatory targets of Med9 and Med10 are involved mostly in amino acid biosynthesis. This result suggests that different sets of Mediator proteins may function in distinct transcriptional regulation pathways. In order to test this hypothesis, we compared the effects of the transcriptional defects of Mediator mutants (med9 null, med10 ts, med11 ts, gal11 null, and med6 ts) on transcription of the GAL1, MFα1, and HIS4 genes, which are regulated specifically by distinct groups of gene-specific transcriptional activator proteins (Fig. 5). Transcription of GAL1 and MFα1 is regulated by Gal4 and Mcm1/Matα1, respectively, whereas regulation of HIS4 transcription requires the concerted participation of Bas1/Bas2, Rap1, and Gcn4 (6). In an amino acid-rich environment, Bas1/Bas2 and Gcn4 are required independently for the basal expression of HIS4, and Rap1 augments DNA binding by Bas1/Bas2 and Gcn4 (6). Upon amino acid starvation, Gcn4 is overproduced by a translational control mechanism and drives Gcn4-mediated transcriptional activation (10).
FIG. 5.
Activator-specific transcriptional defects of Mediator mutants. Poly(A)+ RNA was isolated from wild-type strains (W), mutant strains (M), and mutant strains transformed with the corresponding wild-type Mediator gene (rescued [R]) for each of the Mediator mutants (MED9, MED10, MED11, GAL11, and MED6) noted at the top. In order to measure the levels of the transcripts indicated at the left, cells were grown under galactose induction (GAL1), amino acid-rich conditions (HIS4 B), or amino acid starvation conditions (HIS4 A). All of the yeast cells used in this experiment are α mating type cells and thus display activated MFα1 transcription (MFα1). The levels of MFα1 transcripts in the med9 and med11 mutant strains containing then wild-type Mediator genes (MED9 R and MED11 R) and in the gal11 mutant strain (GAL11 W and M) were not determined. Each blot was hybridized with the probes indicated at the left. For each Northern blot, the level of actin mRNA was measured as an RNA loading control, and a typical result is shown.
Northern analysis revealed that the med9 and med11 mutations reduced specifically basal transcription of HIS4 (an 8-fold reduction) and activated transcription of MFα1 (a 2.3-fold reduction), respectively (Fig. 5, MED9 and MED11). The med10 mutation had no effect on GAL1 transcription but reduced the levels of activated MFα1 transcription (9-fold), as well as basal (5-fold) and activated (15-fold) HIS4 transcription (Fig. 5, MED10). As shown previously (8, 24, 31), the gal11 null and med6 ts mutations diminished severely transcription of GAL1 and MFα1. However, these mutations affected HIS4 transcription in distinct ways. Both basal and activated HIS4 transcription were decreased slightly in the gal11 mutant but were diminished severely in the med6 ts mutant (Fig. 5, GAL11 and MED6). Taken together, these results reveal several interesting phenomena. First, each Mediator mutant exhibited a distinct transcriptional defect pattern. Even the med9 and med10 mutants had completely different effects on HIS4 transcription under amino acid starvation conditions. Second, the fact that Med6 is absolutely required for most of the transcriptional activation events tested suggests that Med6 may function downstream of the other Mediator proteins to modulate Pol II activity.
Activator specificity of Mediator proteins.
The observation of distinct transcriptional defects for individual Mediator mutants suggests that each gene-specific transcriptional activator protein requires a certain Mediator protein(s) to accomplish the transcriptional activation of a particular gene. Specifically, the distinct requirements for Med9, Med10, and Gal11 in the different types of HIS4 transcriptional regulation suggest that each of these Mediator proteins may interact with one of the three HIS4 transcriptional activators, Bas1/Bas2, Gcn4, and Rap1. Because Gal11 has been suggested to interact with Rap1, we examined the effects of the med9, med10, and gal11 mutations on transcriptional activation of ARG4, which contains only the Bas1/Bas2 and Gcn4 binding sites without the Rap1 binding site. The transcriptional defects of ARG4 in med9 and med10 mutants were identical to those of HIS4 in the same mutants. However, ARG4 transcription was not compromised in the gal11 mutant (data not shown), confirming that Gal11 mediates transcriptional stimulation by Rap1.
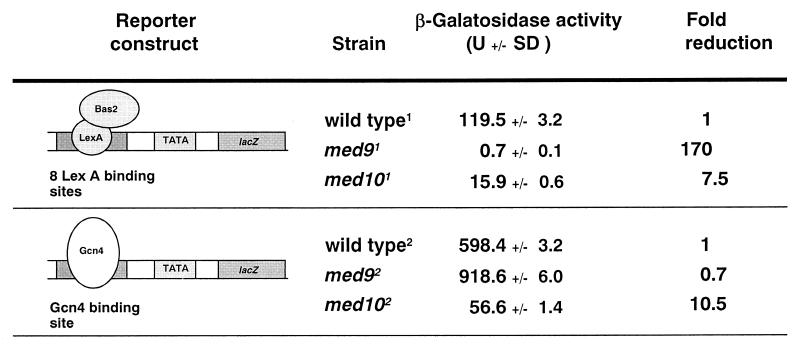
In order to demonstrate that the distinct requirements for Med9 and Med10 in HIS4 transcription resulted from their specific interaction with the Bas1/Bas2 and Gcn4 activators, we used lacZ reporter plasmids bearing either a Gcn4 binding site or multiple LexA binding sites upstream of the CYC1 core promoter. We cotransformed the LexA-driven reporter construct and a multicopy LexA-Bas2 expression construct (43) into wild-type and Mediator mutant strains and examined the levels of lacZ expression at the nonpermissive temperature. Bas2-mediated transcriptional activation was abolished completely in the med9 mutant (a 170-fold reduction) and diminished in the med10 mutant (a 7.5-fold reduction) (Fig. 6). On the other hand, transcriptional activation of the lacZ reporter gene containing the Gcn4 binding site was severely defective only in the med10 mutant when cells were grown under amino acid starvation conditions at the nonpermissive temperature (Fig. 6) (a 10-fold reduction). These results demonstrate that Med9 is required specifically for Bas2-mediated transcriptional activation, whereas Med10 is required for Bas2- and Gcn4-mediated transcriptional activation.
FIG. 6.
Activator-specific requirement for Med9 and Med10. The structures of the lacZ reporter constructs containing a binding site(s) for one activator type are shown along with the transcriptional activator used for the assay. The β-galactosidase activities from the averages of two triplicate assays are shown with standard deviations (SD). The strains used were YPHBAS (wild type1), YSJ9BAS med9 null (_med9_1), and YSJ10BAS med10 ts (_med10_1) for the Bas2 assay, while YPHG4 (wild type2), YSJ9G4 med9 null (_med9_2), and YSJ10G4 med10 ts (_med10_2) were used for the Gcn4 assay.
In vitro transcription of mutant holopolymerase.
The in vivo analysis described above showed that specific activator proteins require a distinct Mediator protein(s) to regulate the Pol II transcription machinery. In order to examine whether the activator-specific properties of Mediator are present in the absence of additional cofactors, we examined the effects of Mediator mutations on basal and activated transcription in a defined in vitro transcription system reconstituted with pure basal transcription factors, activator protein, and holopolymerase.
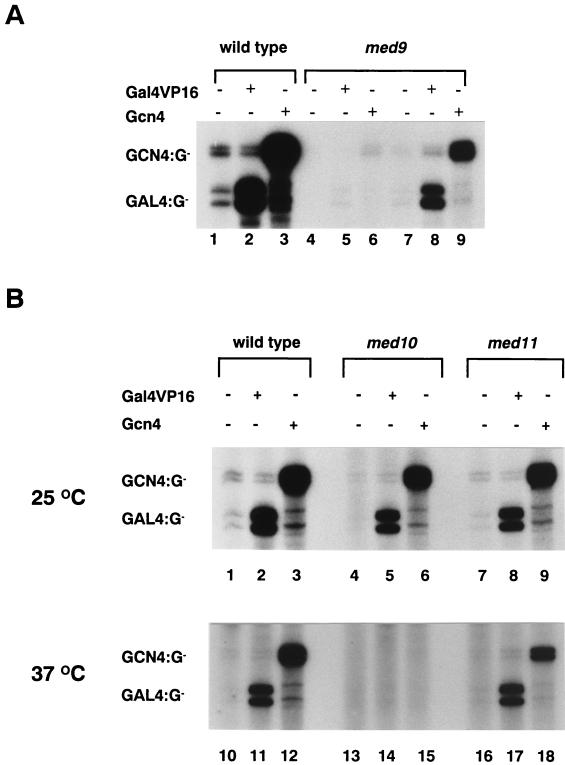
When equivalent amounts (based on nonspecific polymerase activities) of the wild-type and med9 null holopolymerases were tested for basal transcription, the wild-type holopolymerase displayed an eightfold-higher level of basal transcription than did the med9 null holopolymerase (Fig. 7A, lanes 1 to 3 versus lanes 4 to 6). Even when equivalent amounts of Mediator proteins were present, the med9 mutant holopolymerase displayed about 50% of the level of basal transcription activity shown by the wild-type holopolymerase (Fig. 7A, lanes 7 to 9). However, upon addition of the activators Gal4VP16 and Gcn4, both the wild-type (20- to 27-fold activation) and med9 mutant (20- to 28-fold activation) holopolymerases were able to activate transcription from specific enhancer-containing templates (Fig. 7A). This result indicates that Med9 is not required for Gal4VP16- and Gcn4-mediated transcriptional activation.
FIG. 7.
In vitro transcription activities of Mediator mutant holopolymerases. In vitro transcription reactions were reconstituted as described by Lee et al. (24). Activators (Gal4VP16 [30 ng] and Gcn4 [30 ng]) were added to the reaction mixtures as indicated. The reaction mixtures were incubated on ice (A) or at the indicated temperature (B) for 10 min for initiation complex formation, and then [α-32P]UTP (10 μCi) and 0.5 mM CTP were added and the reaction mixtures were incubated further at 25°C for 30 min. Specifically initiated transcripts from a template containing either a Gcn4 binding site (GCN4:G-) or a Gal4 binding site (GAL:G-) are indicated. (A) Transcriptional activity of wild-type holopolymerase (lanes 1 to 3) and equivalent amounts of med9 null holopolymerase based on nonspecific RNA polymerase activity (lanes 4 to 6) or based on Mediator content (lanes 7 to 9). (B) Transcriptional activity of wild-type (lanes 1 to 3 and 10 to 12), med10 ts (lanes 4 to 6 and 13 to 15), and med11 ts (lanes 7 to 9 and 16 to 18) holopolymerases under permissive (25°C, lanes 1 to 9) or restrictive (37°C, lanes 10 to 18) conditions.
In contrast, when equal amounts of wild-type, med10 ts, and med11 ts mutant holopolymerases were tested in vitro, an equivalent level of basal and activated (25- to 30-fold) transcription was observed at the permissive temperature (Fig. 7B, lanes 1 to 9). However, under nonpermissive conditions, the med10 mutant holopolymerase was defective for activated transcription (Fig. 7B, lanes 13 to 15). Although heat treatment reduced significantly the basal activities of the holopolymerases, small amounts of transcripts were detected in the transcription reactions of both wild-type and mutant holopolymerases. However, the robust transcriptional activation of the med10 ts holopolymerase observed under permissive conditions was abolished completely under nonpermissive conditions (Fig. 7B, lanes 5 and 6 versus lanes 14 and 15). Therefore, mediation of the transcriptional activator signals to Pol II was inhibited specifically in the med10 mutant. The fact that both the in vivo and in vitro transcription assays displayed similar defects suggests that Med10 is needed for direct regulation of holopolymerase by specific activator proteins. However, whether Gcn4 and Gal4VP16 utilize identical activation mechanisms or require additional cofactors for activator specificity in vivo is not yet known.
DISCUSSION
Activator-specific subunits of Mediator.
The modular organization of Mediator was first suggested by the identification of the Gal11 module (26). Subsequently, differential salt dissociation experiments (23) and partial reconstitution of the Srb4 Mediator subcomplex with recombinant proteins (19) revealed the presence of the Rgr1 and Srb4 subcomplexes, which appear to function at each end of the signal transfer between transcriptional activators and Pol II (23). Recently, we found that acidic activators bind strongly to the Gal11 protein of the Rgr1 subcomplex, demonstrating that the Gal11 module is an activator binding target (25). However, the gal11 mutation exerts its effect on the transcription of only a limited number of genes (8, 31). This observation suggests that multiple modules or individual Mediator proteins with different activator specificities exist in the Mediator complex. Other Mediator components associated with Rgr1 are candidates for such a role, but genetic evidence for these putative functional interactions was heretofore unavailable. Therefore, the identification of the activator-specific requirements of Med9, Med10, and Med11 supports the hypothesis that the Mediator complex consists of multiple activator-specific components.
It should be noted that the Med6 protein in the Srb4 subcomplex is required for the transcriptional activation of a broader range of genes than are the other Med proteins in the Rgr1 subcomplex. The med6 mutation caused defects in the transcriptional activation of all genes whose transcription is dependent on other specific Mediator proteins. This result suggests that transcriptional activation signals targeted to a specific subunit(s) of Mediator complex may all converge on Med6 during the regulatory process. Med6 may then, in turn, relay the signals to Pol II via the Srb proteins. The general requirement of Srb4 for Pol II transcription suggests that Srb4 functions in the enhancement of basal transcription by Pol II rather than in the mediation of gene-specific activator signals. However, a weak binding affinity between Srb4 and gene-specific transcriptional activators was detected in an in vitro biochemical assay (19). Determination of whether Srb4 constitutes a major target of transcriptional activators in vivo and forms an additional activator-specific module is left for more rigorous examinations.
Activator specificity of the Mediator modules.
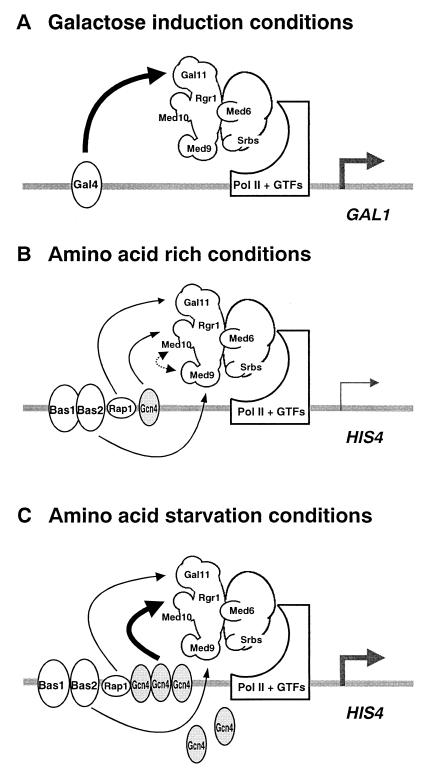
Our results show that deletion of the med9 gene caused a specific defect in Bas1/Bas2-dependent basal HIS4 transcription, while a med10 mutation yielded defects in Gcn4-dependent basal and activated transcription of HIS4. Neither of these mutations had an effect on the transcription of GAL1, which is regulated by the Gal4 protein. Genetic mutations in Gal11 module components cause severe transcriptional defects of Gal4-dependent genes. Thus, the Med9, Med10, and Gal11 modules appear to mediate transcriptional regulatory signals from the Bas1/Bas2, Gcn4, and Gal4 transcriptional activators, respectively (Fig. 8).
FIG. 8.
A model for activator-specific modules of Mediator complex. Three models for activator-specific interactions of the Mediator complex are shown. Filled and linear arrows indicate the specific functional interactions revealed by in vivo transcription assays. Shaded arrows represent transcription initiation. (A) Galactose induction conditions. Gal4 bound to the enhancer interacts with the Gal11 module of Mediator, which induces Pol II and the general transcription factors (GTFs) to transcribe GAL1 at higher efficiency. (B) Amino acid-rich conditions. Med9 and Med10 mediate specifically basal-level HIS4 transcription via Bas1/Bas2 and Gcn4, respectively. Stimulation of Bas1/Bas2- and Gcn4-dependent transcription by Rap1 through the Gal11 module is indicated by a linear arrow. The putative requirement for Med10 in Bas2-mediated transcription is indicated by a dotted arrow. (C) Amino acid starvation conditions. The major transcriptional activation of HIS4 by the specific interaction of (overproduced) Gcn4 protein with Med10 is shown as a filled arrow, compared to the relatively minor contribution from Rap1 to the Gcn4-mediated transcriptional activation via the Gal11 module (linear arrow).
Although Gal4 is not implicated in HIS4 transcription, the level of HIS4 transcription in wild-type yeast strains is two- to threefold higher than that in strains that carry a defective Gal11 module component (gal11 or sin4) (16, 31). However, the transcriptional defects of gal11 and sin4 mutants appear to be related to Rap1 rather than to Bas1/Bas2 or Gcn4. Rap1 binds to the HIS4 promoter and stimulates both Bas1/Bas2- and Gcn4-dependent HIS4 transcription two- to threefold, especially under uninduced conditions (6). Although a direct interaction between the Gal11 module and Rap1 has not been established, sin4 and gal11 mutants have been shown to be defective in the transcription of genes that contain a Rap1 binding site in their promoters (for example, HIS4, PYK1, CTS1, MATα, and Ty1) (16, 31). Genes whose transcription is controlled by Bas1/Bas2 and Gcn4 but which do not have a Rap1 binding site in their promoters (for example, the purine biosynthetic genes and HIS3) are not affected by sin4 or gal11 mutations. In particular, ARG4, which does not have a Rap1 binding site, displays a transcriptional requirement for Med9 and Med10 identical to that of HIS4. However, unlike HIS4, ARG4 does not require the Gal11 module for full-level transcription (data not shown). These observations are consistent with the notion that each Mediator module has a distinct activator specificity.
Future directions.
The identification of the entire complement of Mediator proteins and their functional analyses presented here address one of the major questions with respect to transcriptional activation mechanisms: how is the activator-specific property of Mediator achieved? In addition to the Gal11 module, which is required for Gal4-mediated transcriptional activation, the identification of Med9, Med10, and Med11, which are required for the transcription of distinct groups of genes, clearly demonstrates the existence of activator-specific Mediator pathways. In addition, a group of Mediator subunits, including Rgr1, Sin4, and Gal11, is involved in transcriptional repression as well as activation (8, 15, 16). The observation that the various Mediator functions (activation, repression, and stimulated basal transcription) require different sets of Mediator proteins further supports the notion of multiple functional modules of the Mediator complex. The reconstitution of activator- or repressor-specific Mediator modules with recombinant proteins in vitro should provide a complete biochemical description of the molecular interactions among transcriptional activators and Mediator components.
ACKNOWLEDGMENTS
We thank Jin Mo Park and Juri Kim for technical help, Kelly LaMarco for careful reading of the manuscript, and R. Robinson, D. Kirby, K. Pierce, and E. Spooner of the Harvard Microchemistry Facility for their expertise and technical assistance. We also thank C. Gustafsson, R. Roeder, D. Reinberg, R. Kornberg, and I. Herskowitz for sharing information before publication. We give special thanks to C. Gustafsson, S. Bjoklund, and A. Hinnebusch for Mediator antibodies and Bas2-related plasmids.
This work was supported by grants from SBRI (B-96-004) and the Ministry of Health and Welfare, Republic of Korea (HMP-97-B-3-0030 of the 1997 Good Health R&D project), to Y.-J.K.
REFERENCES
- 1.Apone L M, Virbasius C M, Reese J C, Green M R. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 2.Bjorklund S, Kim Y-J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 3.Brown M T, Goetsch L, Hartwell L H. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen X H, Xiao Z, Fitzgerald-Hayes M. SCM2, a tryptophan permease in Saccharomyces cerevisiae, is important for cell growth. Mol Gen Genet. 1994;244:260–268. doi: 10.1007/BF00285453. [DOI] [PubMed] [Google Scholar]
- 6.Devlin C, Tice-Baldwin K, Shore D, Arndt K T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991;11:3462–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eng J K, McCormick A L, Yates J R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 8.Fassler J S, Winston F. The Saccharomyces cerevisiae SPT13/GAL11 gene has both positive and negative regulatory roles in transcription. Mol Cell Biol. 1989;9:5602–5609. doi: 10.1128/mcb.9.12.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gustafsson C M, Myers L C, Li Y, Redd M J, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. Identification of Rox3 as a component of mediator and RNA polymerase II holoenzyme. J Biol Chem. 1997;272:48–50. doi: 10.1074/jbc.272.1.48. [DOI] [PubMed] [Google Scholar]
- 10.Hinnebusch A G. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch A G, Lucchini G, Fink G R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci USA. 1985;82:498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann A, Horikoshi M, Wang C K, Schroeder S, Weil P A, Roeder R G. Cloning of the Schizosaccharomyces pombe TFIID gene reveals a strong conservation of functional domains present in Saccharomyces cerevisiae TFIID. Genes Dev. 1990;4:1141–1148. doi: 10.1101/gad.4.7.1141. [DOI] [PubMed] [Google Scholar]
- 13.Hoyt M A, Macke J P, Roberts B T, Geiser J R. Saccharomyces cerevisiae PAC2 functions with CIN1, 2 and 4 in a pathway leading to normal microtubule stability. Genetics. 1997;146:849–857. doi: 10.1093/genetics/146.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter T, Plowman G D. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Y W, Dohrmann P R, Stillman D J. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y W, Stillman D J. Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics. 1995;140:103–114. doi: 10.1093/genetics/140.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser K, Meisterernst M. The human general co-factors. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- 18.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 19.Koh S S, Ansari A Z, Ptashne M, Young R A. An activator target in the RNA polymerase II holoenzyme. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 20.Kouprina N, Kirillov A, Kroll E, Koryabin M, Shestopalov B, Bannikov V, Zakharyev V, Larionov V. Identification and cloning of the CHL4 gene controlling chromosome segregation in yeast. Genetics. 1993;135:327–341. doi: 10.1093/genetics/135.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee D K, Wang K C, Roeder R G. Functional significance of the TATA element major groove in transcription initiation by RNA polymerase II. Nucleic Acids Res. 1997;25:4338–4345. doi: 10.1093/nar/25.21.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee T I, Wyrick J J, Koh S S, Jennings E G, Gadbois E L, Young R A. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y C, Kim Y-J. Requirement for a functional interaction between mediator componets Med6 and Srb4 in RNA polymerase II transcription. Mol Cell Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y C, Min S, Gim B S, Kim Y-J. A transcriptional mediator protein that is required for activation of many RNA polymerase II promoters and is conserved from yeast to humans. Mol Cell Biol. 1997;17:4622–4632. doi: 10.1128/mcb.17.8.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, Y. C., J. M. Park, S. Min, S. J. Han, and Y.-J. Kim. An activator binding module of yeast RNA polymerase II holoenzyme. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 26.Li Y, Bjorklund S, Jiang Y W, Kim Y-J, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellor J, Rathjen J, Jiang W, Barnes C A, Dowell S J. DNA binding of CPF1 is required for optimal centromere function but not for maintaining methionine prototrophy in yeast. Nucleic Acids Res. 1991;19:2961–2969. doi: 10.1093/nar/19.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 29.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nash H M, Bruner S D, Scharer O D, Kawate T, Addona T A, Spooner E, Lane W S, Verdine G L. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 31.Nishizawa M, Suzuki Y, Nogi Y, Matsumoto K, Fukasawa T. Yeast Gal11 protein mediates the transcriptional activation signal of two different transacting factors, Gal4 and general regulatory factor I/repressor/activator site binding protein 1/translation upstream factor. Proc Natl Acad Sci USA. 1990;87:5373–5377. doi: 10.1073/pnas.87.14.5373. . (Erratum, 90:302, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piruat J I, Chavez S, Aguilera A. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics. 1997;147:1585–1594. doi: 10.1093/genetics/147.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pugh B F, Tjian R. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J Biol Chem. 1992;267:679–682. [PubMed] [Google Scholar]
- 34.Roeder, R. G. 1998. Personal communication.
- 35.Ruppert S, Tjian R. Human TAFII250 interacts with RAP74: implications for RNA polymerase II initiation. Genes Dev. 1995;9:2747–2755. doi: 10.1101/gad.9.22.2747. [DOI] [PubMed] [Google Scholar]
- 36.Sun X, Zhang Y, Cho H, Ricket P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki Y, Nogi Y, Abe A, Fukasawa T. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4991–4999. doi: 10.1128/mcb.8.11.4991. . (Erratum, 12:4806, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabtiang R K, Herskowitz I. Nuclear proteins Nut1p and Nut2p cooperate to negatively regulate a Swi4p-dependent lacZ reporter gene in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4707–4718. doi: 10.1128/mcb.18.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 40.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 41.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIS. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 42.Xiao Z, McGrew J T, Schroeder A J, Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang F, Kirouac M, Zhu N, Hinnebusch A G, Rolfes R J. Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene transcription. Mol Cell Biol. 1997;17:3272–3283. doi: 10.1128/mcb.17.6.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]