Mining SNPs from EST databases - PubMed (original) (raw)
Mining SNPs from EST databases
L Picoult-Newberg et al. Genome Res. 1999 Feb.
Abstract
There is considerable interest in the discovery and characterization of single nucleotide polymorphisms (SNPs) to enable the analysis of the potential relationships between human genotype and phenotype. Here we present a strategy that permits the rapid discovery of SNPs from publicly available expressed sequence tag (EST) databases. From a set of ESTs derived from 19 different cDNA libraries, we assembled 300,000 distinct sequences and identified 850 mismatches from contiguous EST data sets (candidate SNP sites), without de novo sequencing. Through a polymerase-mediated, single-base, primer extension technique, Genetic Bit Analysis (GBA), we confirmed the presence of a subset of these candidate SNP sites and have estimated the allele frequencies in three human populations with different ethnic origins. Altogether, our approach provides a basis for rapid and efficient regional and genome-wide SNP discovery using data assembled from sequences from different libraries of cDNAs.
Figures
Figure 1
An example of a contig containing a high quality mismatch. (a) A Consed view of a contig containing sequences from the 3′-untranslated region of the erythroblastosis virus oncogene homolog 2 (ETS2). The arrow indicates the location of the high-quality mismatch (A vs. T, position 207). Examples of sequence traces show an A at position 207 (b) and a T at position 207 (c). The mismatch has been confirmed as a common SNP by DNA sequence analysis.
Figure 2
Examples of two SNPs in the GADD34 gene confirmed by DNA sequence analysis. (a) Homozygote C vs. heterozygote C/T; (b) homozygote G vs. heterozygote G/C. Less than 2% of contigs contained more than one SNP. When confirmed the majority of these were in complete linkage disequilibrium with one another.
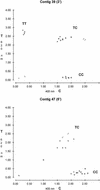
Figure 3
Cluster analyses of candidate EST–SNP sites of GBA genotype data from two contigs. The contig number and library source (3′ or 5′) are given. Each GBA experiment assayed 18 genomic DNA samples from three ethnic groups: eight Caucasian DNA samples (+), five African American DNA samples (o), and five Hispanic DNA samples (x), as well as two positive (/) and two negative controls (\ , −). Raw optical density values from the two-color ELISA system were plotted on an xy scatterplot. Allele 1 data (fluorescein–PNPP reactions) were captured by a standard plate reader at 405 nm and were plotted on the _x_-axis. Allele 2 readings (biotin–TMB reactions, 620 nm) were plotted on the _y_-axis. Thus, the axes represent the base analog extension signals determined by primer extension. Homozygotes for allele 1 lie on the _x_-axis, homozygotes for allele 2 lie on the _y_-axis, and heterozygotes lie on the diagonal. Negative PCR controls (\, reactions controlling for cross-hybridization of the PCR primer and capture–extension primer) and negative GBA controls (−, reactions controlling for self-extension of the capture–extension primer) lie near the plot origins. Synthetic templates (/) were also included as positive controls to monitor hybridization and extension efficiency. Cluster analyses for genotype determination were done automatically by an in-house software program.
Similar articles
- Mining SNPs from EST sequences using filters and ensemble classifiers.
Wang J, Zou Q, Guo MZ. Wang J, et al. Genet Mol Res. 2010 May 4;9(2):820-34. doi: 10.4238/vol9-2gmr765. Genet Mol Res. 2010. PMID: 20449815 - Single nucleotide polymorphism hunting in cyberspace.
Gu Z, Hillier L, Kwok PY. Gu Z, et al. Hum Mutat. 1998;12(4):221-5. doi: 10.1002/(SICI)1098-1004(1998)12:4<221::AID-HUMU1>3.0.CO;2-I. Hum Mutat. 1998. PMID: 9744471 Review. - SNP mining porcine ESTs with MAVIANT, a novel tool for SNP evaluation and annotation.
Panitz F, Stengaard H, Hornshøj H, Gorodkin J, Hedegaard J, Cirera S, Thomsen B, Madsen LB, Høj A, Vingborg RK, Zahn B, Wang X, Wang X, Wernersson R, Jørgensen CB, Scheibye-Knudsen K, Arvin T, Lumholdt S, Sawera M, Green T, Nielsen BJ, Havgaard JH, Brunak S, Fredholm M, Bendixen C. Panitz F, et al. Bioinformatics. 2007 Jul 1;23(13):i387-91. doi: 10.1093/bioinformatics/btm192. Bioinformatics. 2007. PMID: 17646321 - Using mtDNA sequences to estimate SNP parameters in ESTs.
Reed KM. Reed KM. Anim Biotechnol. 2008;19(3):166-77. doi: 10.1080/10495390802170916. Anim Biotechnol. 2008. PMID: 18607789 - The cDNA sequencing project.
Urushihara H, Morio T, Tanaka Y. Urushihara H, et al. Methods Mol Biol. 2006;346:31-49. doi: 10.1385/1-59745-144-4:31. Methods Mol Biol. 2006. PMID: 16957283 Review.
Cited by
- Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data.
Batley J, Barker G, O'Sullivan H, Edwards KJ, Edwards D. Batley J, et al. Plant Physiol. 2003 May;132(1):84-91. doi: 10.1104/pp.102.019422. Plant Physiol. 2003. PMID: 12746514 Free PMC article. - Development of reference transcriptomes for the major field insect pests of cowpea: a toolbox for insect pest management approaches in west Africa.
Agunbiade TA, Sun W, Coates BS, Djouaka R, Tamò M, Ba MN, Binso-Dabire C, Baoua I, Olds BP, Pittendrigh BR. Agunbiade TA, et al. PLoS One. 2013 Nov 22;8(11):e79929. doi: 10.1371/journal.pone.0079929. eCollection 2013. PLoS One. 2013. PMID: 24278221 Free PMC article. - Single nucleotide polymorphism discovery from expressed sequence tags in the waterflea Daphnia magna.
Orsini L, Jansen M, Souche EL, Geldof S, De Meester L. Orsini L, et al. BMC Genomics. 2011 Jun 13;12:309. doi: 10.1186/1471-2164-12-309. BMC Genomics. 2011. PMID: 21668940 Free PMC article. - Mining for single nucleotide polymorphisms in pig genome sequence data.
Kerstens HH, Kollers S, Kommadath A, Del Rosario M, Dibbits B, Kinders SM, Crooijmans RP, Groenen MA. Kerstens HH, et al. BMC Genomics. 2009 Jan 6;10:4. doi: 10.1186/1471-2164-10-4. BMC Genomics. 2009. PMID: 19126189 Free PMC article. - Large-Scale Transcriptome Analysis in Faba Bean (Vicia faba L.) under Ascochyta fabae Infection.
Ocaña S, Seoane P, Bautista R, Palomino C, Claros GM, Torres AM, Madrid E. Ocaña S, et al. PLoS One. 2015 Aug 12;10(8):e0135143. doi: 10.1371/journal.pone.0135143. eCollection 2015. PLoS One. 2015. PMID: 26267359 Free PMC article.
References
- Adams MD, Soares MB, Kerlavage AR, Fields C, Venter JC. Rapid cDNA sequencing (expressed sequence tags) from a directionally cloned human infant brain cDNA library. Nat Genet. 1993;4:373–380. - PubMed
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SP. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. - PubMed
- Cl’ement K, Vaisse C, Lahlow N, Cabrol S, Pelloux V, Cassuto D, Gourmelen M, Dina C, Chambaz J, Lacorte JM, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials