Immunochemical detection and isolation of DNA from metabolically active bacteria - PubMed (original) (raw)
Immunochemical detection and isolation of DNA from metabolically active bacteria
E Urbach et al. Appl Environ Microbiol. 1999 Mar.
Abstract
Most techniques used to assay the growth of microbes in natural communities provide no information on the relationship between microbial productivity and community structure. To identify actively growing bacteria, we adapted a technique from immunocytochemistry to detect and selectively isolate DNA from bacteria incorporating bromodeoxyuridine (BrdU), a thymidine analog. In addition, we developed an immunocytochemical protocol to visualize BrdU-labeled microbial cells. Cultured bacteria and natural populations of aquatic bacterioplankton were pulse-labeled with exogenously supplied BrdU. Incorporation of BrdU into microbial DNA was demonstrated in DNA dot blots probed with anti-BrdU monoclonal antibodies and either peroxidase- or Texas red-conjugated secondary antibodies. BrdU-containing DNA was physically separated from unlabeled DNA by using antibody-coated paramagnetic beads, and the identities of bacteria contributing to both purified, BrdU-containing fractions and unfractionated, starting-material DNAs were determined by length heterogeneity PCR (LH-PCR) analysis. BrdU-containing DNA purified from a mixture of DNAs from labeled and unlabeled cultures showed >90-fold enrichment for the labeled bacterial taxon. The LH-PCR profile for BrdU-containing DNA from a labeled, natural microbial community differed from the profile for the community as a whole, demonstrating that BrdU was incorporated by a taxonomic subset of the community. Immunocytochemical detection of cells with BrdU-labeled DNA was accomplished by in situ probing with anti-BrdU monoclonal antibodies and Texas red-labeled secondary antibodies. Using this suite of techniques, microbial cells incorporating BrdU into their newly synthesized DNA can be quantified and the identities of these actively growing cells can be compared to the composition of the microbial community as a whole. Since not all strains tested could incorporate BrdU, these methods may be most useful when used to gain an understanding of the activities of specific species in the context of their microbial community.
Figures
FIG. 1
Growth kinetics for bacterial cultures with and without BrdU. Arrows indicate times at which cultures were supplemented with BrdU and TdR (+BrdU) or TdR alone (−BrdU). Cultures were harvested at the final time points and used to prepare DNA for dot blots.
FIG. 2
Immunochemical detection of BrdU in DNA dot blots. (A) DNA from cultured bacteria grown in the presence (+) or absence (−) of BrdU, probed with anti-BrdU monoclonal antibodies and peroxidase-conjugated secondary antibodies. (B) DNA from Cronemiller Lake bacterioplankton, incubated with or without BrdU, probed with anti-BrdU monoclonal antibodies and Texas red-conjugated secondary antibodies.
FIG. 3
Immunocytochemical detection of BrdU-labeled Alteromonas sp. strain C250.5-4. The panels on the left show fluorescence from Texas red-conjugated secondary antibodies bound to anti-BrdU monoclonal antibodies; the panels on the right show DAPI fluorescence. The upper panels show BrdU-labeled cultures; the lower panels show unlabeled cultures.
FIG. 4
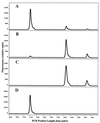
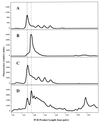
Immunochemical purification of BrdU-containing DNA, analyzed by LH-PCR. (A) Experimental mixture containing DNA from a BrdU-labeled Alteromonas cultures and an unlabeled Roseobacter culture. (B) DNA immunochemically purified from the mixture by using anti-BrdU monoclonal antibodies and paramagnetic beads. (C) BrdU-labeled Alteromonas DNA. (D) Unlabeled Roseobacter DNA. The electropherogram for unlabeled Alteromonas DNA was identical to that shown in panel C.
FIG. 5
LH-PCR analysis of active microbial taxa in Cronemiller Lake. (A) BrdU-labeled lake water DNA. (B) Active fraction isolated from labeled lake water DNA by using anti-BrdU monoclonal antibodies and paramagnetic beads. (C) Control (unlabeled lake water DNA). (D) Control fraction isolated from unlabeled lake water DNA by using anti-BrdU monoclonal antibodies and paramagnetic beads. Dashed lines indicate the positions of the major peaks at 317 and 319 nucleotides.
Similar articles
- Pushing the limits for amplifying BrdU-labeled DNA encoding 16S rRNA: DNA polymerase as the determining factor.
Roux-Michollet DD, Schimel JP, Holden PA. Roux-Michollet DD, et al. J Microbiol Methods. 2010 Dec;83(3):312-6. doi: 10.1016/j.mimet.2010.09.016. Epub 2010 Sep 29. J Microbiol Methods. 2010. PMID: 20883730 - Persistence and growth of fecal Bacteroidales assessed by bromodeoxyuridine immunocapture.
Walters SP, Field KG. Walters SP, et al. Appl Environ Microbiol. 2006 Jul;72(7):4532-9. doi: 10.1128/AEM.00038-06. Appl Environ Microbiol. 2006. PMID: 16820440 Free PMC article. - A survey of the methods for the characterization of microbial consortia and communities.
Spiegelman D, Whissell G, Greer CW. Spiegelman D, et al. Can J Microbiol. 2005 May;51(5):355-86. doi: 10.1139/w05-003. Can J Microbiol. 2005. PMID: 16088332 Review. - Development and application of new nucleic acid-based technologies for microbial community analyses in foods.
Rudi K, Nogva HK, Moen B, Nissen H, Bredholt S, Møretrø T, Naterstad K, Holck A. Rudi K, et al. Int J Food Microbiol. 2002 Sep 15;78(1-2):171-80. doi: 10.1016/s0168-1605(02)00236-2. Int J Food Microbiol. 2002. PMID: 12222632 Review.
Cited by
- Conventional tobacco products harbor unique and heterogenous microbiomes.
Chattopadhyay S, Ramachandran P, Malayil L, Mongodin EF, Sapkota AR. Chattopadhyay S, et al. Environ Res. 2023 Mar 1;220:115205. doi: 10.1016/j.envres.2022.115205. Epub 2022 Dec 30. Environ Res. 2023. PMID: 36592812 Free PMC article. - Cross-kingdom interactions and functional patterns of active microbiota matter in governing deadwood decay.
Purahong W, Tanunchai B, Muszynski S, Maurer F, Wahdan SFM, Malter J, Buscot F, Noll M. Purahong W, et al. Proc Biol Sci. 2022 May 11;289(1974):20220130. doi: 10.1098/rspb.2022.0130. Epub 2022 May 11. Proc Biol Sci. 2022. PMID: 35538788 Free PMC article. - Dynamics of actively dividing prokaryotes in the western Mediterranean Sea.
Mena C, Reglero P, Balbín R, Martín M, Santiago R, Sintes E. Mena C, et al. Sci Rep. 2022 Feb 8;12(1):2064. doi: 10.1038/s41598-022-06120-y. Sci Rep. 2022. PMID: 35136122 Free PMC article. - Coupled DNA-labeling and sequencing approach enables the detection of viable-but-non-culturable Vibrio spp. in irrigation water sources in the Chesapeake Bay watershed.
Malayil L, Chattopadhyay S, Mongodin EF, Sapkota AR. Malayil L, et al. Environ Microbiome. 2021 Jun 22;16(1):13. doi: 10.1186/s40793-021-00382-1. Environ Microbiome. 2021. PMID: 34158117 Free PMC article. - Challenges Faced by Highly Polyploid Bacteria with Limits on DNA Inheritance.
Angert ER. Angert ER. Genome Biol Evol. 2021 Jun 8;13(6):evab037. doi: 10.1093/gbe/evab037. Genome Biol Evol. 2021. PMID: 33677487 Free PMC article. Review.
References
- Ausubel F A, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1988.
- Bard D G, Ward B B. A species-specific bacterial productivity method using immunomagnetic separation and radiotracer experiments. J Microbiol Methods. 1997;28:207–219.
- Brock T D. Bacterial growth rate in the sea: direct analysis by thymidine autoradiography. Science. 1967;155:81–83. - PubMed
- Choi J W, Sherr E B, Sherr B F. Relation between presence-absence of a visible nucleoid and metabolic activity in bacterioplankton cells. Limnol Oceanogr. 1996;41:1161–1168.
- Coote J G, Binnie C. Tolerance to bromodeoxyuridine in a thymidine-requiring strain of Bacillus subtilis. J Gen Microbiol. 1986;132:481–492. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources