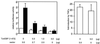Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity - PubMed (original) (raw)
Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity
H Miyakawa et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Hypertonicity (most often present as high salinity) is stressful to the cells of virtually all organisms. Cells survive in a hypertonic environment by increasing the transcription of genes whose products catalyze cellular accumulation of compatible osmolytes. In mammals, the kidney medulla is normally hypertonic because of the urinary concentrating mechanism. Cellular accumulation of compatible osmolytes in the renal medulla is catalyzed by the sodium/myo-inositol cotransporter (SMIT), the sodium/chloride/betaine cotransporter, and aldose reductase (synthesis of sorbitol). The importance of compatible osmolytes is underscored by the necrotic injury of the renal medulla and subsequent renal failure that results from the inhibition of SMIT in vivo by administration of a specific inhibitor. Tonicity-responsive enhancers (TonE) play a key role in hypertonicity-induced transcriptional stimulation of SMIT, sodium/chloride/betaine cotransporter, and aldose reductase. We report the cDNA cloning of human TonE binding protein (TonEBP), a transcription factor that stimulates transcription through its binding to TonE sequences via a Rel-like DNA binding domain. Western blot and immunohistochemical analyses of cells cultured in hypertonic medium reveal that exposure to hypertonicity elicits slow activation of TonEBP, which is the result of an increase in TonEBP amount and translocation to the nucleus.
Figures
Figure 1
TonEBP is a Rel-like protein. (A) The predicted amino acid sequence of human TonEBP. The NFAT-like Rel homology domain and two stretches of glutamine residues are underlined. (B) Sequence alignment of Rel-homology domains in TonEBP and NFAT isoforms (21). Amino acids conserved in all of the members are shaded.
Figure 2
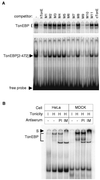
(A) DNA binding profile of the recombinant TonEBP. The N-terminal 472 amino acids of TonEBP (amino acids 2–472—TonEBP[2–472]), including the Rel-like domain, were bacterially expressed and purified. EMSA was performed by using 0.3 nM 32P-hTonE without or with 10 nM competitors as indicated on top: double-stranded DNA probes hTonE, hTonE mutants—M1 to M11—and cTonE were described previously (15). Each binding reaction includes 10 μg of nuclear extract from HeLa cells cultured in hypertonic medium (Upper) or 0.05 μg of TonEBP[2–472] (Lower). The reactions were separated on nondenaturing gels, and autoradiograms of the gels are shown. Only the top portion of the gel is shown for the HeLa nuclear extracts. Bands representing the native TonEBP (from HeLa cells), TonEBP[2–472], and free probe are marked at left. (B) Super-shift analysis of TonEBP. Each lane contains 10 μg of nuclear extract prepared from HeLa or MDCK cells cultured in hypertonic medium for 18 hours (H) or in isotonic medium (I). The antiserum raised against TonEBP[2–472] (IM) and preimmune serum (PI) were added to binding mixtures—0.1 μl or serum added undiluted per reaction—as indicated. The gel was run longer than those in A to allow the super-shifted bands to move into the gel. The TonEBP bands and the super-shifted TonEBP band (S) are marked at left.
Figure 3
TonEBP[1–472] inhibits stimulation of transcription by hypertonicity. (Left) MDCK cells were transiently transfected with 2 μg of a plasmid containing a TonE-driven luciferase gene (15) along with plasmid pcDNA3.1(+) (Invitrogen) directing expression of TonEBP[1–472] or pcDNA3.1(+) by itself (vector) in amounts indicated at the bottom. Transfected cells were cultured in isotonic (open bars) or hypertonic medium (filled bars) for 18 hours, and the luciferase activity was measured. Luciferase activity was normalized to the control cells [transfected with the pcDNA3.1(+) and the luciferase construct] cultured in isotonic medium. Results are mean ± SEM; n = 4. (Right) Lack of effect of TonEBP[1–472] on tumor necrosis factor α-induced NF-κB activity. MDCK cells were transfected with 2 μg of a plasmid containing the luciferase gene under the control of κB sequence from the immunoglobin κ light chain gene (19) along with 3 μg of TonEBP[1–472] in pcDNA3.1(+) or pcDNA3.1(+) by itself as indicated at the bottom. The transfected cells were treated without or with tumor necrosis factor α (20 ng/ml) for 6 hours and were analyzed for luciferase activity. Fold-induction of luciferase by tumor necrosis factor α treatment is shown. Results are mean ± SEM; n = 4.
Figure 4
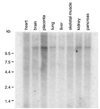
Expression of TonEBP mRNA in human tissues. A blot containing poly(A) RNA from human tissues (CLONTECH) was probed with TonEBP cDNA. All of the tissues shown express TonEBP mRNA. Similar results (not shown) also were obtained with RNA from spleen, thymus, prostate, testis, ovary, small intestine, and colon.
Figure 5
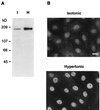
Regulation of TonEBP by hypertonicity. (A) Western blot analysis of whole cell lysates prepared from MDCK cells cultured in isotonic (I) or hypertonic (H) medium for 18 hours by using the antiserum raised against TonEBP[2–472] (Fig. 2). (B) Distribution of TonEBP in isotonic and hypertonic MDCK cells. MDCK cells were grown on glass coverslips and were treated with either isotonic or hypertonic medium for 18 hours. Cells then were fixed and stained with the TonEBP antiserum. (Bar = 20 μm.)
Comment in
- What sets the TonE during osmotic stress?
Kültz D, Csonka L. Kültz D, et al. Proc Natl Acad Sci U S A. 1999 Mar 2;96(5):1814-6. doi: 10.1073/pnas.96.5.1814. Proc Natl Acad Sci U S A. 1999. PMID: 10051549 Free PMC article. Review. No abstract available.
Similar articles
- Bidirectional regulation of tonicity-responsive enhancer binding protein in response to changes in tonicity.
Woo SK, Dahl SC, Handler JS, Kwon HM. Woo SK, et al. Am J Physiol Renal Physiol. 2000 Jun;278(6):F1006-12. doi: 10.1152/ajprenal.2000.278.6.F1006. Am J Physiol Renal Physiol. 2000. PMID: 10836989 - Transcription of the sodium/myo-inositol cotransporter gene is regulated by multiple tonicity-responsive enhancers spread over 50 kilobase pairs in the 5'-flanking region.
Rim JS, Atta MG, Dahl SC, Berry GT, Handler JS, Kwon HM. Rim JS, et al. J Biol Chem. 1998 Aug 7;273(32):20615-21. doi: 10.1074/jbc.273.32.20615. J Biol Chem. 1998. PMID: 9685419 Free PMC article. - Nuclear redistribution of tonicity-responsive enhancer binding protein requires proteasome activity.
Woo SK, Maouyo D, Handler JS, Kwon HM. Woo SK, et al. Am J Physiol Cell Physiol. 2000 Feb;278(2):C323-30. doi: 10.1152/ajpcell.2000.278.2.C323. Am J Physiol Cell Physiol. 2000. PMID: 10666027 - Kidney cell survival in high tonicity.
Handler JS, Kwon HM. Handler JS, et al. Comp Biochem Physiol A Physiol. 1997 Jul;117(3):301-6. doi: 10.1016/s0300-9629(96)00267-8. Comp Biochem Physiol A Physiol. 1997. PMID: 9172386 Review. - Regulation of the myo-inositol and betaine cotransporters by tonicity.
Handler JS, Kwon HM. Handler JS, et al. Kidney Int. 1996 Jun;49(6):1682-3. doi: 10.1038/ki.1996.246. Kidney Int. 1996. PMID: 8743476 Review.
Cited by
- Peptide affinity analysis of proteins that bind to an unstructured region containing the transactivating domain of the osmoprotective transcription factor NFAT5.
Dumond JF, Zhang X, Izumi Y, Ramkissoon K, Wang G, Gucek M, Wang X, Burg MB, Ferraris JD. Dumond JF, et al. Physiol Genomics. 2016 Nov 1;48(11):835-849. doi: 10.1152/physiolgenomics.00100.2016. Epub 2016 Oct 7. Physiol Genomics. 2016. PMID: 27764768 Free PMC article. - Knockdown of Tcirg1 inhibits large-osteoclast generation by down-regulating NFATc1 and IP3R2 expression.
Zhang D, Lin L, Yang B, Meng Z, Zhang B. Zhang D, et al. PLoS One. 2020 Aug 13;15(8):e0237354. doi: 10.1371/journal.pone.0237354. eCollection 2020. PLoS One. 2020. PMID: 32790690 Free PMC article. - Effects of hyperosmolality on expression of urea transporter A2 and aquaporin 2 in mouse medullary collecting duct cells.
Jin W, Yao X, Wang T, Ji Q, Li Y, Yang X, Yao L. Jin W, et al. J Huazhong Univ Sci Technolog Med Sci. 2012 Feb;32(1):59-64. doi: 10.1007/s11596-012-0010-1. Epub 2012 Jan 27. J Huazhong Univ Sci Technolog Med Sci. 2012. PMID: 22282246 - A Gain-of-Function Cleavage of TonEBP by Coronavirus NSP5 to Suppress IFN-β Expression.
Park H, Lee SM, Jeong SJ, Kweon YC, Shin GW, Kim WY, Lee-Kwon W, Park CY, Kwon HM. Park H, et al. Cells. 2024 Sep 26;13(19):1614. doi: 10.3390/cells13191614. Cells. 2024. PMID: 39404379 Free PMC article. - LPS-induced NFκB enhanceosome requires TonEBP/NFAT5 without DNA binding.
Lee HH, Sanada S, An SM, Ye BJ, Lee JH, Seo YK, Lee C, Lee-Kwon W, Küper C, Neuhofer W, Choi SY, Kwon HM. Lee HH, et al. Sci Rep. 2016 Apr 27;6:24921. doi: 10.1038/srep24921. Sci Rep. 2016. PMID: 27118681 Free PMC article.
References
- Yancey P H, Clark M E, Hand S C, Bowlus R D, Somero G N. Science. 1982;217:1214–1222. - PubMed
- Lucht J M, Bremer E. FEMS Microbiol Rev. 1994;14:3–20. - PubMed
- Ishitani M, Nakamura T, Han S Y, Takabe T. Plant Mol Physiol. 1995;27:307–315. - PubMed
- Yamauchi A, Uchida S, Preston A S, Kwon H M, Handler J S. Am J Physiol. 1993;264:F20–F23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases