Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope - PubMed (original) (raw)
Review
Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope
W W Navarre et al. Microbiol Mol Biol Rev. 1999 Mar.
Abstract
The cell wall envelope of gram-positive bacteria is a macromolecular, exoskeletal organelle that is assembled and turned over at designated sites. The cell wall also functions as a surface organelle that allows gram-positive pathogens to interact with their environment, in particular the tissues of the infected host. All of these functions require that surface proteins and enzymes be properly targeted to the cell wall envelope. Two basic mechanisms, cell wall sorting and targeting, have been identified. Cell well sorting is the covalent attachment of surface proteins to the peptidoglycan via a C-terminal sorting signal that contains a consensus LPXTG sequence. More than 100 proteins that possess cell wall-sorting signals, including the M proteins of Streptococcus pyogenes, protein A of Staphylococcus aureus, and several internalins of Listeria monocytogenes, have been identified. Cell wall targeting involves the noncovalent attachment of proteins to the cell surface via specialized binding domains. Several of these wall-binding domains appear to interact with secondary wall polymers that are associated with the peptidoglycan, for example teichoic acids and polysaccharides. Proteins that are targeted to the cell surface include muralytic enzymes such as autolysins, lysostaphin, and phage lytic enzymes. Other examples for targeted proteins are the surface S-layer proteins of bacilli and clostridia, as well as virulence factors required for the pathogenesis of L. monocytogenes (internalin B) and Streptococcus pneumoniae (PspA) infections. In this review we describe the mechanisms for both sorting and targeting of proteins to the envelope of gram-positive bacteria and review the functions of known surface proteins.
Figures
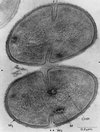
FIG. 1
Transmission electron micrograph of _S. aureus_cells undergoing cell division. Completed division septa are indicated by a single arrowhead, and new division sites are marked by double arrowheads. Newly formed cell wall (W2) is distinguished from preexisting cell wall (W1). Also visible are the bacterial chromosome (Chr) and cytoplasmic membranous bodies (M). This electron micrograph was generated by P. Giesbrecht. Reprinted from reference with permission of the publisher.
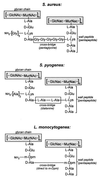
FIG. 2
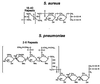
Diagram of peptidoglycan structures from S. aureus, S. pyogenes, and L. monocytogenes. Glycan chains composed of a repeating disaccharide, GlcNAc and MurNAc, are linked to short wall peptides through the lactyl moeity of MurNAc. Adjacent wall peptides can be linked through crossbridge peptides (pentaglycine in S. aureus or dialanine in S. pyogenes). In some cases, such as L. monocytogenes, adjacent wall peptides are not cross-linked via peptide crossbridges. Instead, wall peptides are linked via an amide bond between the ɛ-amino group of a _meso_-diaminopimelic acid (m-Dpm) residue at position 3 of one wall peptide and the carboxy terminus of the
d
-alanyl residue at position 4 of the adjacent wall peptide.
FIG. 3
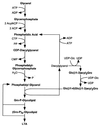
Diagram of the biosynthetic pathway of cell wall assembly. As discussed in the text, the generation of cell wall precursors begins in the cytoplasm, resulting in the synthesis of UDP-MurNAc–
l
-Ala–
d
-iGlu–
l
-Lys–
d
-Ala–
d
-Ala (Park’s nucleotide). The peptidoglycan precursor subunit is transferred to a lipid carrier in the membrane to generate lipid I. After further modification, the lipid-anchored peptidoglycan precursor is translocated to the extracellular face of the cytoplasmic membrane. The peptidoglycan precursor is subsequently incorporated into the cell wall by transpeptidation and transglycosylation reactions with the concomitant displacement of the lipid carrier. The terminal
d
-alanine of the wall pentapeptide is subject to substitution by cross-linking to other wall subunits or can be removed by the action of a
d
-alanyl–
d
-alanine carboxypeptidase. PEP, phosphoenolpyruvate; MN-GN, MurNAc-GlcNAc.
FIG. 4
Structure of cell wall teichoic acids from S. aureus, B. subtilis, and L. monocytogenes. Wall teichoic acids are anionic polymers of glycerol-phosphate, ribitol-phosphate, or glucosyl-phosphate and are tethered to the peptidoglycan. The cell wall linkage unit consists of ManNac(β1-4)GlcNAc, which is 1–6 1-6phosphodiester linked to MurNAc within the glycan strands of the cell wall envelope. One to three glycerol-phosphate moieties serve as a bridge between the linkage unit and the anionic polymer, which is composed of 15 to 60 subunits. R indicates covalent modification of the hydroxyl side chains of either glycerol-phosphate or ribitol-phosphate and can be either GlcNAc or esterified
d
-alanine.
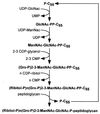
FIG. 5
Diagram of the biosynthesis of staphylococcal cell wall teichoic acid. Synthesis begins in the cytoplasm via the addition of UDP-GlcNAc to an undecaprenol carrier molecule (P-C55). UDP-ManNac and 2,3-CDP-glycerol are added to complete the cell wall linkage unit, which is then extended by the polymerization of ribitol-phosphate. This precursor molecule is thought to be translocated across the cytoplasmic membrane and linked to MurNAc within the glycan chains.
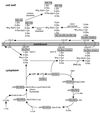
FIG. 6
Structure of lipoteichoic acid from _S. aureus_and S. pneumoniae. Staphylococcal lipoteichoic acid is composed of polymerized glycerol-phosphate and is linked to a membrane anchor moiety [Glc(β1–4)Glc(β1–3)diacylglycerol]. The fatty acid side chains of diacylglycerol are indicated by R. The hydroxyl side chains of polymerized glycerolphosphate are modified with GlcNAc or esterified
d
-alanine. In S. pneumoniae, the repeat unit of lipoteichoic acid, [Glc(β1–3)AATGal(α14)GalNac(6-Cho-P)(α1–3)GalNac(6-Cho-P)(β1-1)ribitol-P], is identical to that of cell wall teichoic acids. The choline-modified repeat unit serves as a targeting receptor for several murein hydrolases and surface protein. Modified from reference with permission of the publisher.
FIG. 7
Diagram of the biosynthesis of staphylococcal lipoteichoic acid (LTA). Glycerol-phosphate is acylated with two fatty acids to yield phosphatidic acid. This compound, as well as all other biosynthetic components of lipoteichoic acid, are thought to be located in the outer leaftlet of the cytoplasmic membrane, the presumed site of lipoteichoic acid synthesis. The glycolipid anchor, Glc (β1–4)Glc(β1–3)diacylglycerol, is linked to the first glycerol-phosphate moiety and then further extended by polymerization with phosphatidyl-glycerol as a substrate. Modified from reference with permission of the publisher.
FIG. 8
Phenotypes of protein A cell wall sorting signal deletion mutants. Full-length protein A is found to be quantitatively anchored to the cell wall. Deletions of the charged tail, charged tail and hydrophobic domain, or the entire sorting signal result in the complete secretion of the polypeptide into the extracellular milieu. Deletion of the LPXTG motif of the C terminal abolishes the anchoring of the polypeptide to the cell wall. In the case of the protein A sorting signal, these mutant proteins are loosely associated with the cytoplasmic membrane.
FIG. 9
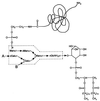
Proposed model for the cell wall sorting reaction. The model can be divided into four distinct steps. 1, the full-length precursor is exported from the cytoplasm via an N-terminal leader peptide. 2, the protein is prevented from release into the extracellular milieu by the charged tail and hydrophobic domain. 3, the protein is cleaved between the threonyl and glycyl residues of the LPXTG motif. 4, the newly liberated carboxy terminus of threonine is transferred via an amide bond exchange to an amino group found at the end of the wall crossbridge. In this proposed model, steps 3 and 4 are coupled in a two-step reaction. Overall, the reaction bears similarity to the amide bond exchange mechanisms by which proteins are attached to GPI moieties and by which transpeptidation occurs during cell wall synthesis.
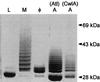
FIG. 10
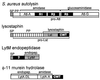
SDS-PAGE of surface protein solubilized with five different muralytic enzymes. Shown is a Coomassie blue-stained SDS-PAGE gel of a hybrid surface protein containing the cell wall sorting signal of protein A after solubilization from the staphylococcal peptidoglycan with lysostaphin (L), mutanolysin (M), the staphylococcal phage φ11 murein hydrolase (φ), the amidase portion of the major staphylococcal autolysin (A, Atl), or the lytic amidase from B. subtilis(A, CwlA). Proteins solubilized with these enzymes display distinct mobilities on SDS-PAGE due to the presence of covalently linked cell wall subunits (see the text). See Fig. 17 for a diagram of the enzymatic activities of these enzymes. A species of 28 kDa is apparent in the amidase-, phage hydrolase-, and mutanolysin-digested samples, which migrates faster than the lysostaphin-solubilized protein. The generation of this species is apparently due to degradation during the prolonged incubation necessary for complete solubilization of the cell wall with these enzymes. Reprinted from reference with permission of the publisher.
FIG. 11
Structure of a surface protein linked to the peptidoglycan of S. aureus. As shown here, the protein is linked to a wall subunit that has been incorporated into the peptidoglycan network by both transpeptidation and transglycosylation. The neighboring wall subunit exists as a pentapeptide and is therefore not substituted. Adapted from reference with permission of the publisher.
FIG. 12
Lipid and peptidoglycan attachment of the murein (Braun’s) lipoprotein of E. coli. The complete processing of the lipoprotein occurs in several discrete steps. 1, The protein is exported from the cytosol by virtue of a type II N-terminal leader peptide. 2, The cysteine residue of the leader peptide is modified by the addition of diacylglycerol. 3, The leader peptide is removed by the type II leader peptidase. 4, The newly liberated amino terminus is acylated. 5, The protein is translocated to the periplasmic side of the outer membrane. 6, The C-terminal lysine residue of the lipoprotein is amide linked to a carboxyl group of the diaminopimelic acid at position 3 of a wall peptide. Like the surface proteins of gram-positive bacteria, the lipoprotein is amide linked to the peptidoglycan. However, in this case the cell wall donates the carboxyl group rather than the amino group to this bond.
FIG. 13
Core structure of a GPI anchor. This diagram shows the core GPI anchor structure found to be common among almost all known GPI anchors from several species. Species-specific variation of the anchor structure can occur through the addition of glycan moieties that can be attached at the points labeled A and B.
FIG. 14
Attachment of a GPI anchor to the C-terminal end of a polypeptide chain. 1, polypeptides entering the secretory pathway are transiently anchored to the membrane of the ER via the hydrophobic portion of the C-terminal GPI-anchoring signal. 2, cleavage occurs between the ω and ω+1 residues. 3, the newly liberated carboxy-terminus of the ω residue is amide linked to the amino group of the ethanolamine of the GPI anchor. This mechanism is similar to the mechanism by which cell wall sorting of surface proteins is proposed to occur in gram-positive bacteria.
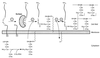
FIG. 15
Diagram of the structures of several anchored surface proteins from various gram-positive bacteria. All surface proteins possess an N-terminal leader peptide (N) and a C-terminal cell wall sorting signal. Many surface proteins possess several repeating domains. Particularly striking are the Cα and Rib proteins of_S. agalactiae_, which are composed of 9 or 12 nearly identical repeat domains, respectively (see the text). References for these proteins are given in Table 1.
FIG. 16
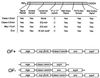
Domain structure of the M-protein family and the structure of the Mga regulons from OF+ and OF−strains of GAS (see the text).
FIG. 17
Sites of hydrolysis of various muralytic enzymes on the peptidoglycan of S. aureus (see the text). Modified from reference with permission of the publisher.
FIG. 18
Domain structure of the peptidoglycan hydrolases of_S. aureus_. Autolysin is synthesized as a preproenzyme, and after cleavage of its N-terminal signal peptide (SP), soluble pro-Atl is released into the extracellular milieu. The repeat domains R1 to R3 function to direct pro-Atl to its receptor on the equatorial surface rings of staphylococci. Proteolytic cleavage generates a propeptide of unknown function (PP), mature amidase with C-terminal R1 and R2 repeats (Atl-A), and glucosaminidase with an N-terminal R3 repeat (Atl-G). Lysostaphin is also synthesized as a preproenzyme, and after signal peptide cleavage, soluble pro-Lst is released into the extracellular milieu. Proteolytic cleavage of 15 tandem repeats of a 13-residue peptide generate mature Lst, which functions as a glycyl-glycine endopeptidase. The C-terminal cell wall-targeting domain (CWT) directs Lst to its receptor on the surface of S. aureus. LytM endopeptidase is probably exported by an N-terminal signal peptide. Its C-terminal domain displays some homology to the endopeptidase domain of lysostaphin, whereas the N-terminal domain of LytM is uncharacterized. φ11 murein hydrolase is encoded by a lysogenic phage (φ11) and exported by a lysis mechanism requiring another phage protein (holein). The enzyme is composed of two domains, one of which is thought to be responsible for amidase activity (LytA-A) and the other is responsible for
d
-Ala-gly endopeptidase activity (LytA-DL). The C-terminal cell wall-targeting signal of LytA displays homology to that of lysostaphin and also functions to direct this enzyme to its receptor in the cell wall envelope of S. aureus.
FIG. 19
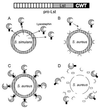
Targeting of lysostaphin to the cell wall envelope of_S. aureus_. (A and B) After secretion by S. simulans bv. staphylolyticus (A), prolysostaphin is converted to the active mature species and targeted to those staphylococci that harbor the appropriate receptor on the bacterial surface (B). (C and D) Mature lysostaphin is a glycyl-glycine endopeptidase that cleaves the peptidoglycan crossbridge of staphylococci (C), thereby hydrolyzing the cell wall envelope and lysing the bacteria (D).
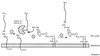
FIG. 20
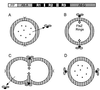
Targeting of Atl to the equatorial surface rings of_S. aureus_. Autolysin is exported from the bacterial cytoplasm (A), and extracellular pro-Atl is targeted to receptor molecules representing the equatorial surface rings of staphylococci that mark the future cell division site (B). Targeting requires the repeats domains (R1 to R3) or pro-Atl. Proteolytic cleavage of pro-Atl generates mature amidase (Atl-A) and glucosaminidase (Atl-G), each of which retains one or two repeat domains and remains bound to the surface rings. The enzymes hydrolyze the cell wall at the cell division site, an event that is probably synchronized with intracellular contraction of FtsZ rings (C), thereby generating two fully separated daughter cells (D). In staphylococci, division occurs perpendicular to the previous cell division plane and targeting of pro-Atl to the second equatorial surface ring can initiate hydrolysis at the future cell division site.
Similar articles
- Structure, function, and assembly of cell walls of gram-positive bacteria.
Shockman GD, Barrett JF. Shockman GD, et al. Annu Rev Microbiol. 1983;37:501-27. doi: 10.1146/annurev.mi.37.100183.002441. Annu Rev Microbiol. 1983. PMID: 6139058 Review. No abstract available. - l-Rhamnosylation of wall teichoic acids promotes efficient surface association of Listeria monocytogenes virulence factors InlB and Ami through interaction with GW domains.
Carvalho F, Sousa S, Cabanes D. Carvalho F, et al. Environ Microbiol. 2018 Nov;20(11):3941-3951. doi: 10.1111/1462-2920.14351. Epub 2018 Jul 31. Environ Microbiol. 2018. PMID: 29984543 - Spatial Organization of Cell Wall-Anchored Proteins at the Surface of Gram-Positive Bacteria.
Dramsi S, Bierne H. Dramsi S, et al. Curr Top Microbiol Immunol. 2017;404:177-201. doi: 10.1007/82_2016_4. Curr Top Microbiol Immunol. 2017. PMID: 27025379 Review. - Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus.
Mazmanian SK, Ton-That H, Schneewind O. Mazmanian SK, et al. Mol Microbiol. 2001 Jun;40(5):1049-57. doi: 10.1046/j.1365-2958.2001.02411.x. Mol Microbiol. 2001. PMID: 11401711 Review. - Envelope Structures of Gram-Positive Bacteria.
Rajagopal M, Walker S. Rajagopal M, et al. Curr Top Microbiol Immunol. 2017;404:1-44. doi: 10.1007/82_2015_5021. Curr Top Microbiol Immunol. 2017. PMID: 26919863 Free PMC article. Review.
Cited by
- Emerging roles of tRNA in adaptive translation, signalling dynamics and disease.
Kirchner S, Ignatova Z. Kirchner S, et al. Nat Rev Genet. 2015 Feb;16(2):98-112. doi: 10.1038/nrg3861. Epub 2014 Dec 23. Nat Rev Genet. 2015. PMID: 25534324 Review. - Two Putative Polysaccharide Deacetylases Are Required for Osmotic Stability and Cell Shape Maintenance in Bacillus anthracis.
Arnaouteli S, Giastas P, Andreou A, Tzanodaskalaki M, Aldridge C, Tzartos SJ, Vollmer W, Eliopoulos E, Bouriotis V. Arnaouteli S, et al. J Biol Chem. 2015 May 22;290(21):13465-78. doi: 10.1074/jbc.M115.640029. Epub 2015 Mar 30. J Biol Chem. 2015. PMID: 25825488 Free PMC article. - Staphylococcus aureus cell wall structure and dynamics during host-pathogen interaction.
Sutton JAF, Carnell OT, Lafage L, Gray J, Biboy J, Gibson JF, Pollitt EJG, Tazoll SC, Turnbull W, Hajdamowicz NH, Salamaga B, Pidwill GR, Condliffe AM, Renshaw SA, Vollmer W, Foster SJ. Sutton JAF, et al. PLoS Pathog. 2021 Mar 31;17(3):e1009468. doi: 10.1371/journal.ppat.1009468. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33788901 Free PMC article. - Biofilms: Formation, drug resistance and alternatives to conventional approaches.
Mirghani R, Saba T, Khaliq H, Mitchell J, Do L, Chambi L, Diaz K, Kennedy T, Alkassab K, Huynh T, Elmi M, Martinez J, Sawan S, Rijal G. Mirghani R, et al. AIMS Microbiol. 2022 Jul 4;8(3):239-277. doi: 10.3934/microbiol.2022019. eCollection 2022. AIMS Microbiol. 2022. PMID: 36317001 Free PMC article. Review. - Chemical Profiling and Antimicrobial Properties of Honey Bee (Apis mellifera L.) Venom.
Tanuwidjaja I, Svečnjak L, Gugić D, Levanić M, Jurić S, Vinceković M, Mrkonjić Fuka M. Tanuwidjaja I, et al. Molecules. 2021 May 20;26(10):3049. doi: 10.3390/molecules26103049. Molecules. 2021. PMID: 34065282 Free PMC article.
References
- Adler L A, Arvidson S. Correlation between the rate of exoprotein synthesis and the amount of the multiprotein complex on membrane-bound ribosomes (MBRP-complex) in Staphylococcus aureus. J Gen Microbiol. 1987;133:803–813. - PubMed
- Åkerström B, Björck L. Protein L: an immunoglobulin light chain-binding bacterial protein. Characterization of binding and physicochemical properties. J Biol Chem. 1989;264:19740–19746. - PubMed
- Åkerström B, Lindqvist A, Lindahl G. Binding properties of protein Arp, a bacterial IgA-receptor. Mol Immunol. 1991;28:349–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI38897/AI/NIAID NIH HHS/United States
- R01 AI038897/AI/NIAID NIH HHS/United States
- T32 GM007185/GM/NIGMS NIH HHS/United States
- GM 07185-22/GM/NIGMS NIH HHS/United States
- AI33985/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases