The DNA helicase activity of BLM is necessary for the correction of the genomic instability of bloom syndrome cells - PubMed (original) (raw)
The DNA helicase activity of BLM is necessary for the correction of the genomic instability of bloom syndrome cells
N F Neff et al. Mol Biol Cell. 1999 Mar.
Abstract
Bloom syndrome (BS) is a rare autosomal recessive disorder characterized by growth deficiency, immunodeficiency, genomic instability, and the early development of cancers of many types. BLM, the protein encoded by BLM, the gene mutated in BS, is localized in nuclear foci and absent from BS cells. BLM encodes a DNA helicase, and proteins from three missense alleles lack displacement activity. BLM transfected into BS cells reduces the frequency of sister chromatid exchanges and restores BLM in the nucleus. Missense alleles fail to reduce the sister chromatid exchanges in transfected BS cells or restore the normal nuclear pattern. BLM complements a phenotype of a Saccharomyces cerevisiae sgs1 top3 strain, and the missense alleles do not. This work demonstrates the importance of the enzymatic activity of BLM for its function and nuclear localization pattern.
Figures
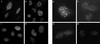
Figure 1
SCE assay. (A) Metaphase chromosomes from SV40-transformed BS fibroblast cell line HG2522 differentially stained to show the high frequency of SCEs. (B) Metaphase chromosomes from SV40-transformed BS fibroblast cell line HG2522 transfected with the normal BLM cDNA. (C) A graphic display of the range and distribution of SCEs in the fibroblast cell lines transfected with cDNAs encoding normal and missense BLM proteins. The mean number of SCEs per 46 chromosomes and range are as follows: HG2522 (55 and 16–88); CAT (55 and 18–105); R12 [WT] (24 and 5–74); Q672R (71 and 38–104); K695T (59 and 33–98); and C1055S (67 and 39–100). The values reported for cell line HG2522 and for the R12-transfected line represent experimental baseline numbers for these cell lines. At least 25 metaphases were counted for each transfected cell line.
Figure 2
Indirect immunofluorescence study of BLM in the nucleus of normal, Bloom syndrome, and SV40-transformed human fibroblast cell lines. All cells are fixed and stained with BLM antibodies, followed by donkey anti-rabbit secondary antibodies conjugated to Texas Red. Cells in A–E are stained with DAPI. Cells in F–H are not stained with DAPI to show diffuse BLM staining. (A) Normal human fibroblasts (HG2619). (B) SV40-transformed normal human fibroblasts (HG2855). (C) BS fibroblasts (HG2940). (D) SV40-transformed BS fibroblasts (HG2522). (E) HG2522 transfected with the normal BLM cDNA. (F) HG2522 transfected with the Q672R cDNA. (G) HG2522 transfected with the C1055S cDNA. (H) HG2522 transfected with the K695T cDNA.
Figure 3
Western transfer analysis of BLM proteins in cell lines and transfected clones. Twenty micrograms of total cell protein were loaded on each lane, and proteins were displayed on a 5% SDS polyacrylamide gel and transferred to a PVDF membrane. Positions of molecular weight markers and BLM are indicated by arrows. Lane 1: SV40-transformed normal fibroblast cell line (HG2855); lane 2: normal fibroblast cell line (HG2619); lane 3: SV40-transformed BS fibroblast cell line (HG2522) transfected with the normal BLM cDNA (R12c41); lane 4: HG2522 transfected with the normal BLM cDNA (R12c45); lane 5: HG2522; lane 6: HG2522 transfected with the Q672R allele; lane 7: HG2522 transfected with the C1055S allele; lane 8: HG2522 transfected with the K695T allele.
Figure 4
Purification and DNA helicase activity of BLM. (A) Isolation of normal and missense proteins of BLM. The left panel (lanes 1–4) contains isolated BLM proteins transferred to a PVDF membrane and reacted with the BLM antisera. Equal amounts of recovered protein are loaded in each lane. The right panel is a silver-stained polyacrylamide gel of the isolated proteins (lanes 5–8). Equal volumes of equivalent fractions from each preparation are loaded to show relative recoveries of the different proteins from yeast strain AMR 61. Molecular weight markers are indicated on the left. Lanes 1 and 5 contain normal BLM; lanes 2 and 6 contain Q672R missense protein; lanes 3 and 7 contain C1055S missense protein; lanes 4 and 8 contain K695T missense protein. (B) Helicase activity of the normal and missense BLM proteins. Displacement activity is measured using a 3′ end-labeled oligonucleotide 54 bases long annealed to ssMp18 DNA. Lanes marked 0 and Δ95–5 are the substrate (no enzyme) and the product (substrate heated at 95°C for 5 min). Each reaction contains 1 ng of recovered protein: lanes 2 and 3, normal BLM; lanes 4 and 5, missense Q672R BLM; lanes 6 and 7 missense C1055S BLM; lanes 8 and 9 missense K695T BLM. (C) Polarity of the BLM helicase. The direction of movement of the BLM helicase was evaluated using a substrate digested with _Pst_I that creates two oligonucleotides of different sizes at opposite ends of the long ssMp18 DNA. In experiment PstA, both ends of the same substrate molecule were labeled (5′ and 3′) before digestion with _Pst_I, and in PstB two different substrate molecules were labeled either at the 5′ or the 3′ end, digested with _Pst_I and equal amounts of each mixed together in the reaction tubes. Assays were quantitated with a Molecular Dynamics PhosphorImager. (D) Comparison of helicase activities of normal and missense BLM proteins. Displacement activity and DNA-dependent ATPase specific activity is shown as a percentage of normal protein activity.
Figure 5
BLM complements a phenotype of a sgs1 top3 S. cerevisiae strain. Yeast strain AMR61 (sgs1 top3) was transformed with plasmids pYES2, pSGS1, and pC4YES3 (normal BLM cDNA under GAL1 control), as well as with missense alleles of BLM under galactose control. Colonies were picked into microtiter wells, replica-plated onto minimal plates with galactose as the carbon source, and incubated at 30°C for 5 d. A comparison of the growth efficiency of these strains on increasing concentrations of hydroxyurea (HU) is shown. Each row displays six colonies from the transformation plates. Row A = +pYES2; row B = +pSGS1; row C = +pC4YES; row E = +pQ672R/BLM; row F = +pC1055S/BLM; row G = +pK695T/BLM.
References
- Bennett R, Sharp J, Wang JC. Purification and characterization of the Sgs1 DNA helicase activity. J Biol Chem. 1998;273:9644–9650. - PubMed
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
- Ellis N. DNA helicases in inherited human disorders. Curr Opin Gen Dev. 1997;7:354–363. - PubMed
- Ellis NA, Groden J, Ye T-Z, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom syndrome gene product is homologous to RecQ helicases. Cell. 1995a;83:655–666. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials