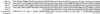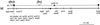Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas - PubMed (original) (raw)
Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas
M E Porter et al. Mol Biol Cell. 1999 Mar.
Abstract
A second cytoplasmic dynein heavy chain (cDhc) has recently been identified in several organisms, and its expression pattern is consistent with a possible role in axoneme assembly. We have used a genetic approach to ask whether cDhc1b is involved in flagellar assembly in Chlamydomonas. Using a modified PCR protocol, we recovered two cDhc sequences distinct from the axonemal Dhc sequences identified previously. cDhc1a is closely related to the major cytoplasmic Dhc, whereas cDhc1b is closely related to the minor cDhc isoform identified in sea urchins, Caenorhabditis elegans, and Tetrahymena. The Chlamydomonas cDhc1b transcript is a low-abundance mRNA whose expression is enhanced by deflagellation. To determine its role in flagellar assembly, we screened a collection of stumpy flagellar (stf) mutants generated by insertional mutagenesis and identified two strains in which portions of the cDhc1b gene have been deleted. The two mutants assemble short flagellar stumps (<1-2 micrometer) filled with aberrant microtubules, raft-like particles, and other amorphous material. The results indicate that cDhc1b is involved in the transport of components required for flagellar assembly in Chlamydomonas.
Figures
Figure 1
Identification of additional Dhc genes in Chlamydomonas. The deduced amino acid sequences of four new Dhc sequences in the region surrounding the conserved ATP hydrolytic site (P-loop 1) are shown. cDhc1a, cDhc1b, and Dhc10 were initially recovered in the RT–PCR screen, whereas Dhc11 was identified during subsequent screening of a genomic library (see MATERIALS AND METHODS for details). Dhc3 is an axonemal Dhc sequence (Porter et al., 1996) that was fortuitously isolated in the PCR screen.
Figure 2
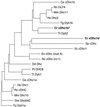
Diagrammatic alignment of cytoplasmic Dhc sequences. The Chlamydomonas reinhardtii (Cr) cytoplasmic Dhc sequences were aligned with cytoplasmic Dhc sequences identified in other organisms using the program CLUSTAL W (Thompson et al., 1994) and were displayed using the program DRAWGRAM from the Phylip package, version 3.57c (Felsenstein, 1998). The abbreviations and GenBank accession numbers for the sequences are as follows: Caenorhabditis elegans (Ce), L33260 and Z75536; Dictyostelium discoideum (Dd), Z15124; Drosophila melanogaster (Dm), L23195; Emericella nidulans (En), U03904; Homo sapiens (Hs), L23958 and U20552; Mus musculus (Mm), Z83808 and Z83809; Neurospora crassa (Nc), L31504; Paramecium tetraurelia (Pt), L17132; Rattus norvegicus (Rn), D13893, L08505, and D26495; Saccharomyces cerevisiae (Sc), Z21877 and L15626; Schizosaccharomyces pombe (Sp), AB006784; Tetrahymena thermophilia (Tt), AF025312 and AF025313; and Tripneustes gratilla (Tg), Z21941 and U03969.
Figure 3
Recovery of the cDhc1b transcription unit. (A) A partial restriction map of the genomic DNA region containing the cDhc1b transcription unit. Also indicated on the diagram are the 150-bp fragment recovered in the PCR screen, the ∼7.7-kb _Sac_I fragment cloned from the size-fractionated minilibrary, the ∼4.6-kb _Not_I fragment used as a Northern probe, and the approximate positions of the seven phage clones obtained from the large insert genomic library. S, _Sac_I sites; N, _Not_I sites. (B) Intron–exon structure of the N-terminal and central region of the cDhc1b gene. A diagram of the relative sizes of the introns and exons is shown. All splice sites were confirmed by RT–PCR. Also indicated are the approximate positions of the TATA box sequences and the proposed translation start site.
Figure 4
Partial amino acid sequence of cDhc1b. The deduced amino acid sequence of the N-terminal and central region of the cDhc1b gene is shown. The four conserved P-loop motifs are indicated in bold letters. EMBL accession number AJ132478.
Figure 5
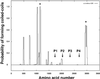
Structural domains within the cDhc1b polypeptide. The probability of forming regions of α-helical coiled-coil structure was determined using the program COILS (Lupus et al., 1991; Lupus, 1996). Peaks of high probability that are also encoded by homologous regions in other Dhc sequences are indicated by the asterisks (see Mitchell and Brown, 1994, 1997).
Figure 6
Pairwise comparison of Dhc sequences. The Chlamydomonas cDhc1b polypeptide was compared with other full-length Dhc sequences using the GCG program Compare with a window size of 50 and a stringency of 22. Shown are the plots against the two C. elegans cytoplasmic dynein sequences, cDhc1a (Lye et al., 1995) and cDhc1b (Wilson et al., 1994). The GenBank accession numbers for the C. elegans sequences are L33260 and Z75536, respectively.
Figure 7
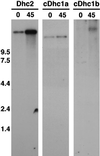
Expression of Chlamydomonas Dhc genes in response to deflagellation. Shown are autoradiograms of Northern blots loaded with total RNA isolated from wild-type cells before (0) and 45 min after (45) deflagellation. Left, the blot was hybridized with a control probe for the axonemal sequence Dhc2 and was exposed overnight. Middle, the blot was hybridized with an ∼6.0-kb _Sac_I fragment from the 5′ end of the cDhc1a transcription unit and was exposed for 20 d, although faint signals could be observed on shorter exposures. Right, the blot was hybridized with a 4.6-kb _Not_I fragment isolated from the 5′ end of the cDhc1b transcription unit (see Figure 3) and was exposed for 10 d. All probes were labeled to the same specific activity, and all of the Dhc transcripts are estimated to be >13 kb in length (see also Porter et al., 1996).
Figure 8
Genetic map position of the cDhc1b gene. The genetic map of linkage group VI (redrawn from Harris, 1989; Porter et al., 1996) is shown on the bottom line. The approximate map locations of the Dhc clones (open triangles) and another molecular marker (black triangle) are shown on the top line. The black squares indicate the genetic markers in the C. reinhardtii strain that were used to anchor the two maps relative to one another. The black circle marks the position of the centromere. The parental ditype:nonparental ditype:tetratype ratios and estimated map distances in centiMorgans (cM) are as follows: cDhc1b versus mt (22:0:1; 2.2 cM), cDhc1b versus Dhc3 (11:0:6; 17.6 cM), cDhc1b versus pcf8–13 (8:0:22; 36.7 cM), and cDhc1b versus act2 (3:0:11; 39.3 cM). The distance from the centromere was estimated using the centromere-linked markers ac17 (4:5:21; 35 cM) and y1 (4:4:20; 35.7 cM). Additional mapping data for the linkage group VI markers are provided in Porter et al. (1996).
Figure 9
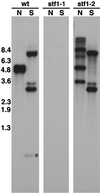
Identification of cDhc1b mutations. Shown are autoradiograms of three duplicate Southern blots containing genomic DNA isolated from wild-type (wt) and two stumpy flagellar mutants, stf1-1 and stf1-2. Four micrograms of genomic DNA were digested with the restriction enzymes _Not_I (N) and _Sac_I (S), separated on a 0.8% agarose gel, transferred to a Magnagraph membrane, and hybridized overnight with the 4.6- and 4.9-kb _Not_I fragments of the cDhc1b gene (see Figure 3). No cross-hybridizing sequence is detected in the stf1-1 genomic DNA, whereas an RFLP in one of the _Not_I fragments is detected in stf1-2 genomic DNA. Longer exposures indicated that the stf1-2 genomic DNA is missing the two _Sac_I fragments of 0.8 and 0.3 kb located within the wild-type 4.6-kb _Not_I fragment (see Figure 3). Hybridization with probes for the NIT1 gene indicated the presence of a single plasmid insert in each strain and the hybridization of this sequence to the polymorphic cDhc1b restriction fragments in stf1-2 (Bower and Porter, unpublished results).
Figure 10
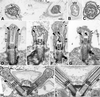
Electron microscopic analysis of stumpy flagellar mutants (A–H) and wild-type Chlamydomonas cells (I). Cross sections of flagella on a single stf1 cell (A) and on individual cells (B and C) typically reveal few complete microtubules and no outer doublet microtubules. Compared with the wild-type flagella (I), the flagella on the stf1 mutants rarely extend beyond the cell wall (D and H). The transition zones and basal bodies appear normal in the mutants, but the flagellar microtubules are short and often terminate as open-ended or filamentous structures (see large arrowheads). Occasionally, microtubules extend from a distal cap-like structure and terminate with the free ends just distal to the basal cup (D, E, and H, small arrowheads). Flagella are filled with electron dense material, and stalked bead structures line the flagellar membrane (small arrows). Bars, 0.1 μm.
Figure 11
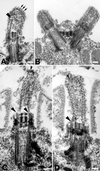
Electron microscopic analysis of detergent-extracted stf1 mutants. Whole cells were extracted with 0.5% Nonidet P-40 for 5 min (A and B) or 2% Nonidet P-40 for 30 min (C and D) before fixation. Both the cell and flagellar membranes have been removed, but amorphous granular and filamentous material remain associated with the flagellar stumps. Filamentous structures extend from the ends of the flagellar or basal body microtubules (A, C, and D, large arrowheads). In some flagella, the filaments associated with the microtubules coalesce at the distal tip (A, small arrows). In others, microtubules with free proximal ends are occasionally found linked to a distal cap-like structure (C, small arrowhead). Bars, 0.1 μm.
Figure 12
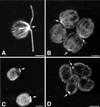
Immunofluorescence microscopy of wild-type (A and C) and stf1 mutant (B and D) cells stained with monoclonal antibodies to β-tubulin (A and B) or one of the kinesin II subunits (C and D). Bars, 2 μm.
Figure 13
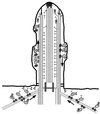
Model for cDhc1b activity in flagellar assembly and maintenance. Transport of IFT particles toward the flagellar tips (the microtubule plus ends) is mediated by FLA10 kinesin II motors (small, paired spheres). Cytoplasmic dynein (large, paired spheres) may be required for the minus-end–directed transport of flagellar components along cytoplasmic microtubules to the base of the flagellum. Alternatively, cytoplasmic dynein may be required to transport IFT particles from the flagellar tip to the flagellar base or for the recycling of IFT particles in the cytoplasm to pick up new components. Although the nature of material transported up the flagellum is unknown, possible components include radial spokes (T-shaped structures), unidentified complexes (large question marks), and/or signaling components involved in regulating flagellar length.
Similar articles
- A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in chlamydomonas and Mammalian cells.
Perrone CA, Tritschler D, Taulman P, Bower R, Yoder BK, Porter ME. Perrone CA, et al. Mol Biol Cell. 2003 May;14(5):2041-56. doi: 10.1091/mbc.e02-10-0682. Epub 2003 Jan 26. Mol Biol Cell. 2003. PMID: 12802074 Free PMC article. - Domains in the 1alpha dynein heavy chain required for inner arm assembly and flagellar motility in Chlamydomonas.
Myster SH, Knott JA, Wysocki KM, O'Toole E, Porter ME. Myster SH, et al. J Cell Biol. 1999 Aug 23;146(4):801-18. doi: 10.1083/jcb.146.4.801. J Cell Biol. 1999. PMID: 10459015 Free PMC article. - The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly.
Pazour GJ, Dickert BL, Witman GB. Pazour GJ, et al. J Cell Biol. 1999 Feb 8;144(3):473-81. doi: 10.1083/jcb.144.3.473. J Cell Biol. 1999. PMID: 9971742 Free PMC article. - Keeping an eye on I1: I1 dynein as a model for flagellar dynein assembly and regulation.
Wirschell M, Hendrickson T, Sale WS. Wirschell M, et al. Cell Motil Cytoskeleton. 2007 Aug;64(8):569-79. doi: 10.1002/cm.20211. Cell Motil Cytoskeleton. 2007. PMID: 17549744 Review. - Axonemal dyneins.
Witman GB. Witman GB. Curr Opin Cell Biol. 1992 Feb;4(1):74-9. doi: 10.1016/0955-0674(92)90061-g. Curr Opin Cell Biol. 1992. PMID: 1532722 Review.
Cited by
- The role of the dynein light intermediate chain in retrograde IFT and flagellar function in Chlamydomonas.
Reck J, Schauer AM, VanderWaal Mills K, Bower R, Tritschler D, Perrone CA, Porter ME. Reck J, et al. Mol Biol Cell. 2016 Aug 1;27(15):2404-22. doi: 10.1091/mbc.E16-03-0191. Epub 2016 Jun 1. Mol Biol Cell. 2016. PMID: 27251063 Free PMC article. - TCTEX1D2 mutations underlie Jeune asphyxiating thoracic dystrophy with impaired retrograde intraflagellar transport.
Schmidts M, Hou Y, Cortés CR, Mans DA, Huber C, Boldt K, Patel M, van Reeuwijk J, Plaza JM, van Beersum SE, Yap ZM, Letteboer SJ, Taylor SP, Herridge W, Johnson CA, Scambler PJ, Ueffing M, Kayserili H, Krakow D, King SM; UK10K; Beales PL, Al-Gazali L, Wicking C, Cormier-Daire V, Roepman R, Mitchison HM, Witman GB. Schmidts M, et al. Nat Commun. 2015 Jun 5;6:7074. doi: 10.1038/ncomms8074. Nat Commun. 2015. PMID: 26044572 Free PMC article. - Label-free proteomic comparison reveals ciliary and nonciliary phenotypes of IFT-A mutants.
Leggere JC, Hibbard JVK, Papoulas O, Lee C, Pearson CG, Marcotte EM, Wallingford JB. Leggere JC, et al. Mol Biol Cell. 2024 Mar 1;35(3):ar39. doi: 10.1091/mbc.E23-03-0084. Epub 2024 Jan 3. Mol Biol Cell. 2024. PMID: 38170584 Free PMC article. - Dyneins across eukaryotes: a comparative genomic analysis.
Wickstead B, Gull K. Wickstead B, et al. Traffic. 2007 Dec;8(12):1708-1721. doi: 10.1111/j.1600-0854.2007.00646.x. Epub 2007 Sep 26. Traffic. 2007. PMID: 17897317 Free PMC article. - Flagellar membrane trafficking in kinetoplastids.
Fridberg A, Buchanan KT, Engman DM. Fridberg A, et al. Parasitol Res. 2007 Jan;100(2):205-12. doi: 10.1007/s00436-006-0329-2. Epub 2006 Oct 21. Parasitol Res. 2007. PMID: 17058110 Review. No abstract available.
References
- Albert PS, Brown SJ, Riddle DL. Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol. 1981;198:435–451. - PubMed
- Asai DJ, Beckwith SM, Kandl KA, Keating HH, Tjandra H, Forney JD. The dynein genes of Paramecium tetraurelia. Sequences adjacent to the catalytic P-loop identify cytoplasmic and axonemal heavy chain isoforms. J Cell Sci. 1994;107:839–847. - PubMed
- Asai DJ, Brokaw CJ. Dynein heavy chain isoforms and axonemal motility. Trends Cell Biol. 1993;3:398–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous