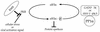A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function - PubMed (original) (raw)
A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function
M Mulvey et al. J Virol. 1999 Apr.
Abstract
The herpes simplex virus type 1 gamma34.5 gene product and the cellular GADD34 protein both contain similar domains that can regulate the activity of eukaryotic initiation factor 2 (eIF2), a critical translation initiation factor. Viral mutants that lack the GADD34-related function grow poorly on a variety of malignant human cells, as activation of the cellular PKR kinase leads to the accumulation of inactive, phosphorylated eIF2 at late times postinfection. Termination of translation prior to the completion of the viral reproductive cycle leads to impaired growth. Extragenic suppressors that regain the ability to synthesize proteins efficiently in the absence of the viral GADD34-related function have been isolated. These suppressor alleles are dominant in trans and affect the steady-state accumulation of several viral mRNA species. We demonstrate that deregulated expression of Us11, a virus-encoded RNA-binding, ribosome-associated protein is necessary and sufficient to confer a growth advantage upon viral mutants that lack a GADD34-related function. Ectopic expression of Us11 reduces the accumulation of the activated cellular PKR kinase and allows for sustained protein synthesis. Thus, an RNA-binding, ribosome-associated protein (Us11) and a GADD34-related protein (gamma34.5) both function in a signal pathway that regulates translation by modulating eIF2 phosphorylation.
Figures
FIG. 1
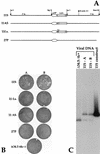
Structure of the HSV-1 SUP locus and summary of mapping data. Deletion plasmids and the extent of the deletions appear below the map. The map represents an enlargement of the Us-TRs junction segment contained in the HSV-1 _Bam_HI Z fragment. Characterized ORFs are represented as open boxes. The Us11 ATG is on the extreme left, while oriS lies on the right. The spliced Us12 transcript and the Us11 transcript (note heterogeneous start sites) appear above the map. Restriction sites are indicated by the arrows over the map. The deletion contained in the SUP1 isolate described by Mohr and Gluzman (35) is shown immediately below the map. The rescue column summarizes data presented in Fig. 2A and B (+, rescues; −, does not rescue).
FIG. 2
Small deletions which cross the boundary between Us and TRs can generate the SUP phenotype. (A) Outline of the marker rescue protocol used in this study. Δ34.5 mutant viral DNA was either transfected alone or cotransfected (+) with a specific rescue plasmid into permissive Vero cells. The rescue plasmids were all WT except for the different internal SUP deletions each contained. After the appearance of CPE, a cell-free lysate was prepared by freeze-thawing, and this lysate was used to infect nonpermissive U373 cells. After passage of the transfection lysate on U373 cells, the viral stock was used to infect freshly confluent 60-mm-diameter dishes of U373 cells and Vero cells. The U373 dishes were fixed, stained, and photographed, while the infected Vero cells were used to isolate viral DNA. ppt., precipitation. (B) After a single passage of the transfection lysates on U373 cells, a cell-free lysate was prepared by freeze-thawing and used to infect a fresh set of confluent U373 cells on 60-mm-diameter dishes. Photographs of these plates, after fixing and staining with crystal violet, are shown. The transfected rescue plasmid appears next to the image of the stained plate. UN, not transfected. (C) Analysis of viral genomes. Lysates from duplicate U373 plates were used to prepare viral DNA in Vero cells. Rescue plasmid DNA clones (C lanes) harboring various deletions within the SUP locus and the corresponding population of rescued viral DNA isolated from Vero cells (V lanes) were digested with _Bam_HI, fractionated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-labeled _Bam_HI-_Bst_E2 DNA fragment that contains the unique portion of the _Bam_HI Z fragment (shown partially in Fig. 1). The filter was washed, and the autoradiogram is shown. As Δ34.5 mutant viruses fail to replicate efficiently on U373 cells, the viral population is enriched with rare recombinants that have acquired a SUP mutation from the targeting plasmid. This Southern analysis demonstrates that viral populations displaying the suppressor phenotype consist of predominantly recombinant viruses which have acquired the genotype specified by the rescue plasmid used in the transfection. The WT _Bam_HI Z fragment of Δ34.5 HSV-1 mutant viruses (WT Z) comigrates with the WT HSV-1 _Bam_HI Z fragment. Δ34.5 viral DNA digested was prepared from stocks propagated only on Vero cells. Additional slow-migrating bands in viral DNA samples (for example, the WT Z fragment in the Δ34.5 mutant) represent expansions within the repetitive TRs regions.
FIG. 3
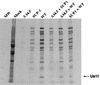
The SUP1 deletion behaves as a _trans_-dominant mutant allele. U373 cells were infected either with each individual virus at an MOI of 5 or infected with a mixture composed of two viruses where each virus was present at an MOI of 5 (the total MOI was 10). Labeled proteins synthesized at late times postinfection were fractionated on SDS-polyacrylamide gels and visualized by autoradiography. MW, molecular weight markers.
FIG. 4
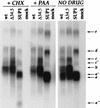
Alterations to the SUP locus affects the steady-state accumulation of multiple RNA species. U373 cells were either infected with each individual virus at high MOI or mock infected in the presence (+) or absence of drug (CHX or PAA). At 6 h postinfection, total RNA was harvested, fractionated by electrophoresis through formaldehyde-agarose gels, and transferred to nylon membranes. To detect transcripts that span the Us-TRs junction, strand-specific RNA probes were prepared from the _Bst_E2-_Xba_I region (see map in Fig. 1). RNAs were detected only with probes antisense to the previously characterized Us11 and Us12 ORFs; furthermore, identical RNAs were observed in WT virus- and Δ34.5-infected cells when an antisense probe within the SUP1 deletion (_Apa_LI-_Eco_NI) was employed (not shown). Approximate sizes of the RNA species (shown to the right of the gel) are as follows: a (Us11), 1.4 kb; b (Us12), 1.9 kb; c, 2.6 kb; d, 3.4 kb; e, 7.9 kb.
FIG. 5
Expression of the Us11 gene product is necessary and sufficient to confer a growth advantage upon Δ34.5 mutants in nonpermissive cells. (A) Schematic of the US11 expression constructs designed to integrate into the HSV-1 tk locus and create tk− recombinants. (B) Fixed plates, stained with crystal violet resulting from a marker rescue experiment. The targeting plasmid used in the transfection appears to the left of the plate. The columns designated A and B refer to two transfections handled independently. (C) Genome analysis of the rescued viral population. A lysate from a duplicate set of U373 plates identical to those shown in panel B was used to infect Vero cells, and viral DNA was isolated. Following digestion with _Eco_RI, DNA was fractionated by agarose gel electrophoresis, blotted onto a nylon membrane, and probed with a fragment from the 3′ tk region. Note that almost all of the DNA in the population, in samples derived from two independent transfections, contains the 11S:tk insertion specified by the 11S targeting construct.
FIG. 6
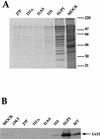
(A) Individual plaque-purified _tk_−:Δ34.5 isolates that express the Us11 polypeptide are capable of enhanced protein synthesis. Nonpermissive U373 cells were infected with the individual viral isolates at high MOI. Proteins labeled in a 1-h pulse with [35S]methionine at 13 h postinfection were fractionated by SDS-PAGE and visualized by autoradiography. The positions of molecular size markers (in kilodaltons) are indicated to the right of the gel. (B) Analysis of steady-state Us11 levels in SUP1 versus Δ34.5 tk−:11S. S10 extracts were prepared from infected U373 cells as described in Materials and Methods. Aliquots were fractionated by SDS-PAGE, electroblotted to Immobilon, and probed with a monoclonal antibody against HSV-1 Us11 (44). Identical results were obtained with extracts prepared directly in 1× Laemmli buffer (not shown).
FIG. 7
Expression of the Us11 polypeptide at immediate-early times precludes the hyperactivation of the cellular PKR kinase. S10 extracts prepared from infected U373 cells were incubated for 30 min at 30°C in the presence of 30 μCi of [γ-32P]ATP. PKR was then immunoprecipitated, and the resulting immune complexes were fractionated by SDS-PAGE and visualized by autoradiography. Bands containing PKR were excised from the gel and counted in liquid scintillant. The positions of molecular size markers (in kilodaltons) are indicated to the left of the gel.
FIG. 8
Us11 inhibits PKR activation and thus allows for sustained translation in the absence of a GADD34-related function. Suppressor viruses display enhanced growth on nonpermissive cells due to their ability to overcome a protein synthesis checkpoint guarded by the cellular PKR kinase. As Us11 is involved in mediating the suppressor phenotype, it could prevent activation of the PKR kinase either by intercepting the activator or inhibiting the activation process (as assessed by autophosphorylation) at a step subsequent to dsRNA binding. P, phosphate; PP1α, protein phosphatase 1α.
Similar articles
- Regulation of eIF2alpha phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle.
Mulvey M, Poppers J, Sternberg D, Mohr I. Mulvey M, et al. J Virol. 2003 Oct;77(20):10917-28. doi: 10.1128/jvi.77.20.10917-10928.2003. J Virol. 2003. PMID: 14512542 Free PMC article. - Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein.
Poppers J, Mulvey M, Khoo D, Mohr I. Poppers J, et al. J Virol. 2000 Dec;74(23):11215-21. doi: 10.1128/jvi.74.23.11215-11221.2000. J Virol. 2000. PMID: 11070019 Free PMC article. - Neutralizing innate host defenses to control viral translation in HSV-1 infected cells.
Mohr I. Mohr I. Int Rev Immunol. 2004 Jan-Apr;23(1-2):199-220. doi: 10.1080/08830180490265600. Int Rev Immunol. 2004. PMID: 14690861 Review.
Cited by
- Host species-specific activity of the poxvirus PKR inhibitors E3 and K3 mediate host range function.
Haller SL, Park C, Bruneau RC, Megawati D, Zhang C, Vipat S, Peng C, Senkevich TG, Brennan G, Tazi L, Rothenburg S. Haller SL, et al. J Virol. 2024 Nov 19;98(11):e0133124. doi: 10.1128/jvi.01331-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480085 Free PMC article. - Biomolecular phase separation in stress granule assembly and virus infection.
Liu Y, Yao Z, Lian G, Yang P. Liu Y, et al. Acta Biochim Biophys Sin (Shanghai). 2023 Jul 3;55(7):1099-1118. doi: 10.3724/abbs.2023117. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37401177 Free PMC article. - Oncolytic herpes simplex viruses for the treatment of glioma and targeting glioblastoma stem-like cells.
Kardani K, Sanchez Gil J, Rabkin SD. Kardani K, et al. Front Cell Infect Microbiol. 2023 May 31;13:1206111. doi: 10.3389/fcimb.2023.1206111. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37325516 Free PMC article. Review. - The Food and Drug Administration-approved antipsychotic drug trifluoperazine, a calmodulin antagonist, inhibits viral replication through PERK-eIF2α axis.
Mao Y, Wang Z, Yao C, Zeng Q, Cheng W, Zhang S, Chen S, Sheng C. Mao Y, et al. Front Microbiol. 2022 Nov 1;13:979904. doi: 10.3389/fmicb.2022.979904. eCollection 2022. Front Microbiol. 2022. PMID: 36386620 Free PMC article. - A little less aggregation a little more replication: Viral manipulation of stress granules.
Brownsword MJ, Locker N. Brownsword MJ, et al. Wiley Interdiscip Rev RNA. 2023 Jan;14(1):e1741. doi: 10.1002/wrna.1741. Epub 2022 Jun 16. Wiley Interdiscip Rev RNA. 2023. PMID: 35709333 Free PMC article. Review.
References
- Brown S M, Harland J. Three mutants of herpes simplex virus type 2: one lacking the genes US10, US11, and US12 and two in which Rs has been extended by 6 Kb to 0.91 map units with loss of Us sequences between 0.94 and the Us/TRs junction. J Gen Virol. 1987;68:1–18. - PubMed
- Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus 1 neurovirulence to γ34.5, a gene nonessential for growth in cell culture. Science. 1990;252:1262–1266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources