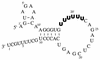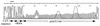Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template - PubMed (original) (raw)
Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template
L L Poon et al. J Virol. 1999 Apr.
Abstract
The poly(A) tail of influenza virus mRNA is thought to be synthesized by reiterative copying of the U track near the 5' end of the virion RNA template. This has been widely accepted as a plausible hypothesis, but until now there has been no direct experimental evidence for it. Here, we report such direct evidence based on the fact that (i) replacing the U track with an A track directs synthesis of products with poly(U) tails, both in vitro and in vivo, and (ii) interrupting the U track abolishes polyadenylation in vitro.
Figures
FIG. 1
vRNA-like template with the wild-type conserved terminal sequences used in the in vitro influenza virus transcription reactions. The Watson-Crick base pairs in the RNA hook model (12) derived from an earlier RNA fork model (2) are shown. The nucleotide numbers starting at the 5′ end are indicated by a prime to distinguish them from nucleotide numbers starting at the 3′ end. The U6 polyadenylation site is shown in bold. (Modified from reference with permission.)
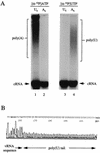
FIG. 2
Mutation of the U6 track to an A6 track of a vRNA-like template. (A) Wild-type vRNA (U6 [lanes 1 and 3]) and mutated RNA (A6 [lanes 2 and 4]) templates are tested in transcription reactions in the presence of [α-32P]ATP (lanes 1 and 2) or [α-32P]UTP (lanes 3 and 4). The cRNA, the polyadenylated mRNA, and the polyuridylated products are indicated. The signal at the origin is thought to be due to transcription products derived from residual endogenous vRNA. When [α-32P]UTP was used in the transcription reaction, the concentration of ATP was increased to 500 μM and the concentration of UTP was reduced to 25 μM. The mobility of the high-molecular-weight smear varies in different gels (lanes 1 and 4). (B) The poly(U) sequence (n = 48) of a cloned RT-PCR product which is derived from the high-molecular-weight transcription product of the A6 mutant (panel A, lane 4). The high-molecular-weight transcription product was eluted from the polyacrylamide gel, reverse transcribed, and amplified by PCR as described previously (11), except that the 5′ GC-clamped T20 primer was replaced by a GC-clamped A20 primer (5′-GCCCCGGGATCCA20-3′).
FIG. 3
Effect of interrupting the U track of the vRNA on polyadenylation activity in vitro. (A) The U6 track is interrupted by inserting a nucleotide (A, C, or G) in the middle. Lane 1, wild-type (WT) RNA (U6); lanes 2 to 4, RNA mutants; lane 5, no template. (B) RNA templates with a single point mutation (U→A) within the U6 track. Lane 1, wild-type (WT) RNA; lanes 2 to 7, point mutants; lane 8, no template. For the numbering scheme, see Fig. 1.
FIG. 4
The poly(U) sequence of a cloned RT-PCR product derived from the poly(U)-tailed CAT mRNA (n = 67). The CAT sequence, vRNA, and poly(U) tail are indicated, with a line drawn over the TAA translation terminator of CAT. Two broad artifact dye blobs centered on residues 82 and 97, respectively, overlap part of the poly(U) sequence. Plasmids pGT-h-PB1, pGT-h-PB2, pGT-h-PA, and pGT-h-NP, which express the PB1, PB2, PA, and NP proteins, respectively, under the control of the adenovirus 2 major late promoter, and pPOLI-CAT-RT (see the text) were generously supplied by Peter Palese. One microgram of each of the pGT-h-PB1, pGT-h-PB2, pGT-h-PA, and pGT-h-NP plasmids and the mutated pPOLI-CAT-RT plasmids were transfected into 293 cells in 30-mm-diameter dishes with 25 μl of DOTAP transfection reagent (Boehringer Mannheim). At 36 h posttransfection, RNA was isolated with TRIZOL reagent (Life Technologies). RNA (3 μg) was reverse transcribed, amplified by PCR, and cloned as described previously (11), except that the 5′ GC-clamped T20 primer was replaced by a GC-clamped A20 primer.
Similar articles
- Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5' conserved sequences.
Pritlove DC, Poon LL, Fodor E, Sharps J, Brownlee GG. Pritlove DC, et al. J Virol. 1998 Feb;72(2):1280-6. doi: 10.1128/JVI.72.2.1280-1286.1998. J Virol. 1998. PMID: 9445028 Free PMC article. - Polyuridylated mRNA synthesized by a recombinant influenza virus is defective in nuclear export.
Poon LL, Fodor E, Brownlee GG. Poon LL, et al. J Virol. 2000 Jan;74(1):418-27. doi: 10.1128/jvi.74.1.418-427.2000. J Virol. 2000. PMID: 10590131 Free PMC article. - Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer.
Shapiro GI, Krug RM. Shapiro GI, et al. J Virol. 1988 Jul;62(7):2285-90. doi: 10.1128/JVI.62.7.2285-2290.1988. J Virol. 1988. PMID: 2453679 Free PMC article. - Transcription and replication of the influenza a virus genome.
Mikulásová A, Varecková E, Fodor E. Mikulásová A, et al. Acta Virol. 2000 Oct;44(5):273-82. Acta Virol. 2000. PMID: 11252672 Review. - Influenza virus transcription.
Skehel JJ, Hay AJ. Skehel JJ, et al. J Gen Virol. 1978 Apr;39(1):1-8. doi: 10.1099/0022-1317-39-1-1. J Gen Virol. 1978. PMID: 347032 Review. No abstract available.
Cited by
- Efficient genome replication in influenza A virus requires NS2 and sequence beyond the canonical promoter.
Swaminath S, Mendes M, Zhang Y, Remick KA, Mejia I, Güereca M, Te Velthuis AJW, Russell AB. Swaminath S, et al. bioRxiv [Preprint]. 2024 Sep 10:2024.09.10.612348. doi: 10.1101/2024.09.10.612348. bioRxiv. 2024. PMID: 39314307 Free PMC article. Preprint. - Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation.
Koppstein D, Ashour J, Bartel DP. Koppstein D, et al. Nucleic Acids Res. 2015 May 26;43(10):5052-64. doi: 10.1093/nar/gkv333. Epub 2015 Apr 21. Nucleic Acids Res. 2015. PMID: 25901029 Free PMC article. - Modulation of mRNA 3'-End Processing and Transcription Termination in Virus-Infected Cells.
Vijayakumar A, Park A, Steitz JA. Vijayakumar A, et al. Front Immunol. 2022 Feb 10;13:828665. doi: 10.3389/fimmu.2022.828665. eCollection 2022. Front Immunol. 2022. PMID: 35222412 Free PMC article. Review. - The Influenza Virus RNA-Polymerase and the Host RNA-Polymerase II: RPB4 Is Targeted by a PB2 Domain That Is Involved in Viral Transcription.
Morel J, Sedano L, Lejal N, Da Costa B, Batsché E, Muchardt C, Delmas B. Morel J, et al. Viruses. 2022 Mar 3;14(3):518. doi: 10.3390/v14030518. Viruses. 2022. PMID: 35336925 Free PMC article. - Type B and type A influenza polymerases have evolved distinct binding interfaces to recruit the RNA polymerase II CTD.
Krischuns T, Isel C, Drncova P, Lukarska M, Pflug A, Paisant S, Navratil V, Cusack S, Naffakh N. Krischuns T, et al. PLoS Pathog. 2022 May 23;18(5):e1010328. doi: 10.1371/journal.ppat.1010328. eCollection 2022 May. PLoS Pathog. 2022. PMID: 35605026 Free PMC article.
References
- Desselberger U, Racaniello V R, Zazra J J, Palese P. The 3′ and 5′ terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. - PubMed
- Hay A J, Lomniczi B, Bellamy A R, Skehel J J. Transcription of the influenza virus genome. Virology. 1977;83:337–355. - PubMed
- Hay A J, Skehel J J, McCauley J. Characterization of influenza virus RNA complete transcripts. Virology. 1982;116:517–522. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources