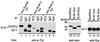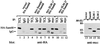Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis - PubMed (original) (raw)
Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis
M Di Fruscio et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Sam68, the 68-kDa Src substrate associated during mitosis, is an RNA-binding protein with signaling properties that contains a GSG (GRP33, Sam68, GLD-1) domain. Here we report the cloning of two Sam68-like-mammalian proteins, SLM-1 and SLM-2. These proteins have an approximately 70% sequence identity with Sam68 in their GSG domain. SLM-1 and SLM-2 have the characteristic Sam68 SH2 and SH3 domain binding sites. SLM-1 is an RNA-binding protein that is tyrosine phosphorylated by Src during mitosis. SLM-1 bound the SH2 and SH3 domains of p59(fyn), Grb-2, phospholipase Cgamma-1 (PLCgamma-1), and/or p120(rasGAP), suggesting it may function as a multifunctional adapter protein for Src during mitosis. SLM-2 is an RNA-binding protein that is not tyrosine phosphorylated by Src or p59(fyn). Moreover, SLM-2 did not associate with the SH3 domains of p59(fyn), Grb-2, PLCgamma-1, or p120(rasGAP), suggesting that SLM-2 may not function as an adapter protein for these proteins. The identification of SLM-1 and SLM-2 demonstrates the presence of a Sam68/SLM family whose members have the potential to link signaling pathways with RNA metabolism.
Figures
Figure 1
Amino acid sequence comparison of mouse SLM-1, SLM-2, and Sam68. The mouse Sam68 sequence is shown on top in capital letters. The mouse SLM-1 and SLM-2 amino acid sequences are shown below. The GSG domain is boxed and the KH domain is marked by a line above the sequence. A hyphen in the sequence denotes a gap. The consensus is shown below and a capital letter denotes sequence identity among all three proteins, a lowercase letter represents sequence identity in two sequences, and a dot represents no sequence identity. The SLM-1 proline-rich sequences are labeled P1 to P4 and the proline motif of SLM-2 is labeled P1. The YDN sequence is underlined and denotes a Grb-2 SH2 domain-binding consensus sequence.
Figure 2
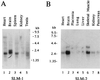
Northern blot analysis of SLM-1 and SLM-2. (A) The mouse multiple tissue Northern blot membrane was hybridized with an SLM-1 DNA probe. (B) The human multiple tissue Northern blot membrane was hybridized, using EST 530290 as a DNA probe. Reprobing of the membranes with Sam68 and/or actin DNA probes showed equal loading.
Figure 3
SLM-1, but not SLM-2, is a substrate for the Src kinase p59fyn. Sam68 (lanes 1–3), SLM-1 (lanes 4–6), and SLM-2 (lanes 7–9) were co-expressed with p59fyn in HeLa cells. The cells were lysed and proteins were immunoprecipitated with IgG or anti-myc antibodies. The bound proteins were separated by SDS/PAGE and immunoblotted with anti-phosphotyrosine antibodies. The migration of Sam68, SLM-1, and the antibody heavy chain are shown. The total cell lysates were also immunoblotted with anti-myc (lanes 10–12) and anti-p59fyn (lanes 13–15) antibodies to verify protein expression.
Figure 4
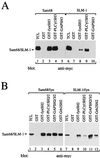
SLM-1 associates with the p59fyn and PLCγ-1 SH3 domains and the p59fyn, PLCγ-1, GAP, and Grb2 SH2 domains. (A) myc-Sam68 and myc-SLM-1 were transfected in HeLa cells separately. The cell lysates were divided equally and incubated with affinity matrices containing GST alone, GST-fynSH3, GST-PLCγ-1SH3, or GST-GAPSH3. The bound proteins were identified by immunoblotting with anti-myc antibodies. The migration position of Sam68 and SLM-1 is shown. (B) myc-Sam68 and myc-SLM-1 were cotransfected with p59fyn. The cell lysates were divided equally and incubated with affinity matrices containing GST alone, GST-fynSH2, GST-PLCγ-1SH2/2, GST-GAP2/3/2, or GST-Grb2SH2. The bound proteins were identified by immunoblotting with anti-myc antibodies.
Figure 5
SLM-1 and SLM-2 bind homopolymeric RNA with different specificities. HeLa cells transfected with myc-SLM-1 (A) or myc-SLM-2 (B) were lysed and incubated with poly(A), poly(C), poly(G), or poly(U) covalently coupled to beads in the absence of competitor (lanes 1–6) or in the presence of 10 or 100 μg of soluble homopolymeric RNA as indicated (lanes 7–16). The bound SLM-1 or SLM-2 was detected by immunoblotting with anti-myc antibodies. TCL, total cell lysate; C, control Sepharose beads.
Figure 6
SLM-1 and SLM-2 associate with Sam68. Vector alone, myc-SLM-1, myc-SLM-2, and myc-Sam68 were cotransfected with HA-Sam68 in HeLa cells as indicated. The transfected cells were lysed and the lysates were immunoprecipitated with IgG or anti-myc antibodies. The immunoprecipitated proteins (IP) were separated by SDS/PAGE and immunoblotted for the presence of HA-Sam68 with anti-HA antibodies. The migration positions of HA-Sam68 and the heavy chain of IgG are shown on the left. The expression of the myc-Sam68, -SLM-1, and -SLM-2 is shown to be equivalent (lanes 14–16).
Figure 7
SLM-1 is a substrate for Src during mitosis. (A) v-Src-transformed cells transfected with vector alone, GFP-Sam68, GFP-SLM-1, or GFP-SLM-2 were treated overnight with 40 ng/ml nocodazole. The cells were lysed and immunoprecipitated with normal rabbit serum (NRS; lanes 1, 3, 5, and 7) or the C20 anti-Sam68 antibody (lanes 2, 4, 6, and 8), which recognizes Sam68, SLM-1, and SLM-2. The immunoprecipitated proteins were separated by SDS/PAGE, transferred to nitrocellulose, and immunoblotted with anti-phosphotyrosine antibodies. (B) The expression of GFP-Sam68, GFP-SLM-1, and GFP-SLM-2 in A is shown by blotting with C20 anti-Sam68. The positions of molecular mass markers are shown on the left in kDa. The positions of GFP-Sam68, GFP-SLM-1, and endogenous Sam68 are shown.
References
- Fumagalli S, Totty N F, Hsuan J J, Courtneidge S A. Nature (London) 1994;368:871–874. - PubMed
- Taylor S J, Shalloway D. Nature (London) 1994;368:867–871. - PubMed
- Vogel L B, Fujita D J. J Biol Chem. 1995;270:2506–2511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous