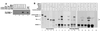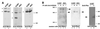HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells - PubMed (original) (raw)
HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells
W F Shen et al. Mol Cell Biol. 1999 Apr.
Abstract
Aberrant activation of the HOX, MEIS, and PBX homeodomain protein families is associated with leukemias, and retrovirally driven coexpression of HOXA9 and MEIS1 is sufficient to induce myeloid leukemia in mice. Previous studies have demonstrated that HOX-9 and HOX-10 paralog proteins are unique among HOX homeodomain proteins in their capacity to form in vitro cooperative DNA binding complexes with either the PBX or MEIS protein. Furthermore, PBX and MEIS proteins have been shown to form in vivo heterodimeric DNA binding complexes with each other. We now show that in vitro DNA site selection for MEIS1 in the presence of HOXA9 and PBX yields a consensus PBX-HOXA9 site. MEIS1 enhances in vitro HOXA9-PBX protein complex formation in the absence of DNA and forms a trimeric electrophoretic mobility shift assay (EMSA) complex with these proteins on an oligonucleotide containing a PBX-HOXA9 site. Myeloid cell nuclear extracts produce EMSA complexes which appear to contain HOXA9, PBX2, and MEIS1, while immunoprecipitation of HOXA9 from these extracts results in coprecipitation of PBX2 and MEIS1. In myeloid cells, HOXA9, MEIS1, and PBX2 are all strongly expressed in the nucleus, where a portion of their signals are colocalized within nuclear speckles. However, cotransfection of HOXA9 and PBX2 with or without MEIS1 minimally influences transcription of a reporter gene containing multiple PBX-HOXA9 binding sites. Taken together, these data suggest that in myeloid leukemia cells MEIS1 forms trimeric complexes with PBX and HOXA9, which in turn can bind to consensus PBX-HOXA9 DNA targets.
Figures
FIG. 1
MEIS1 forms protein-protein complexes with HOXA9 and PBX1a in the presence and absence of DNA. (A) Immunoprecipitation of target DNA complexed to PBX and HOXA9 proteins is mediated by MEIS1 protein. To demonstrate ternary complex formation on a DNA target, an epitope-tagged MEIS1 protein was mixed with untagged HOXA9 and PBX1a proteins and a 32P-labeled DNA target containing a PBX-HOXA9 consensus binding site. Immunoprecipitation with antiserum to the MEIS1 epitope tag brought down the target DNA (lane 3). The DNA was not precipitated when either the PBX and HOXA9 proteins (lane 1) or the MEIS1 protein (lane 2) was omitted. Lane 4 shows the migration position of the input labeled oligonucleotide. (B) MEIS1 forms a triple complex with PBX and HOXA9 in the absence of DNA. [35S]methionine-labeled, FLAG epitope-tagged HOXA9 protein and untagged PBX1a, PBX2, or PBX3 protein, along with a control luciferase protein, were synthesized in vitro, treated with DNase to remove contaminating DNA, and incubated with or without labeled untagged MEIS1b protein. Lanes 1 to 6 show the migration positions of the input proteins. Lanes 7 through 17 were incubated with antiserum to the epitope tag on the HOXA9 protein to precipitate HOXA9 and associated proteins. Immunoprecipitation of the epitope-tagged HOXA9 protein in the absence of MEIS1b resulted in precipitation of low amounts of PBX1a (lane 7), PBX2 (lane 9), or PBX3 (lane 11). Addition of MEIS1b resulted in a 5- to 10-fold increase in precipitation of the PBX1a protein (lane 8), a 7-fold increase in PBX2 (lane 10), and a 3-fold increase in PBX3 (lane 12). The control protein was not precipitated, demonstrating that the protein-protein interactions were specific. To ensure that contaminating DNAs were not facilitating the observed protein-protein interactions, immunoprecipitation experiments were performed in the presence of EtdBr to prevent protein-DNA interactions (43) (lanes 13 to 18). With EtdBr present, addition of MEIS1b resulted in a 10-fold enhancement of PBX2-HOXA9 binding (lanes 13 and 14). In a similar manner, the binding of MEIS1 to HOXA9 was increased fivefold in the presence of PBX2 (compare lane 15 with lane 14). Lanes 16 and 17 are a longer autoradiographic exposure of lanes 14 and 15, to more clearly demonstrate the previously reported observation that MEIS1 binds to HOXA9 in the absence of PBX proteins (41). The MEIS1b protein reproducibly migrated as a diffuse band in the presence of EtdBr (lanes 14 to 17). When either the epitope-specific antibody (lane 18) or the epitope-tagged HOXA9 protein (not shown) was omitted from the reaction mixture, no proteins were precipitated. The brackets mark the position of the various PBX protein bands coprecipitated with HOXA9.
FIG. 2
Formation of a MEIS1-PBX1a-HOXA9 ternary complex on a PBX-HOXA9 DNA target. EMSA was used to assess DNA binding by in vitro-translated proteins. As previously described, PBX1a and HOXA9 form a dimeric binding complex on their consensus target (lane 7), while HOXA9 alone forms a faster-migrating EMSA band (lane 3). Comparison with a lysate control (lane 1) demonstrates that neither PBX1a alone (lane 2), MEIS1b alone (lane 4), MEIS1b with HOXA9 (lane 5), nor MEIS1b with PBX1a (lane 6) is capable of binding this oligonucleotide. Addition of increasing amounts of MEIS1b to PBX1a and HOXA9 resulted in the formation of a new, slower-migrating band and the disappearance of the dimer band (lanes 8 to 12). Preincubation of the triple protein mixture with antiserum to the epitope tag fused to the MEIS1 protein prevented formation of the slower-migrating band together with the appearance of a weak supershifted band and the reappearance of the dimer band (lane 13). The amount of MEIS1 used in lane 13 was the same as that used in lane 9. We interpret this result to mean that the MEIS1 antiserum supershifts some complex while preventing most of the MEIS1b protein from binding to the PBX-HOXA9 dimeric DNA gel shift complex.
FIG. 3
Coimmunoprecipitation of HOXA9 and MEIS or PBX from myeloid cell nuclear extracts. (A) HOXA9, MEIS1, and PBX2 are expressed in U-937 and KG1 nuclear cell extracts. Western gel analysis of U-937 and KG1 cell nuclear extracts using affinity-purified specific antibodies showed expression of MEIS1 (lanes 1 and 2), HOXA9 (lanes 3 and 4), and PBX2 (lanes 5 and 6). (B) Coimmunoprecipitation of HOXA9 with PBX2 or HOXA9 with MEIS1. U-937 and KG1 cell nuclear proteins were incubated with affinity-purified chicken antibody to HOXA9 (lanes 2, 3, 6, and 7) or nonimmune IgY (lanes 1, 4, 5, and 8). Following immunoprecipitation, the pellets were assayed by Western blotting using rabbit antibodies to MEIS1 proteins (lanes 1 to 4) or to PBX2 protein (lanes 5 to 8). Immunoprecipitation of HOXA9 brought down MEIS1 (lanes 2 and 3) and PBX2 (lanes 6 and 7). In a similar manner, U-937 cell nuclear proteins were subjected to immunoprecipitation with a mixture of whole rabbit antisera to PBX protein and affinity-purified antibodies to PBX1, PBX2, and PBX3 (lane 10), or control sera (lane 9), and the pellets were analyzed by Western blotting with affinity-purified chicken antibodies to HOXA9 (lanes 9 and 10). Specific HOXA9 bands were detected in proteins precipitated with anti-PBX (lane 10), while immunoprecipitation with a nonspecific serum did not yield HOXA9 immunoreactive protein (lane 9).
FIG. 4
MEIS1, PBX, and HOXA9 proteins in U-937 cell nuclear extracts can form a ternary EMSA complex. Increasing amounts of U-937 cell nuclear extract shifted an oligonucleotide (oligo) containing the PBX-HOXA9 consensus binding site to form a highly reproducible upper band and a variable lower band (lanes 1 to 3). Both bands could be competed with cold specific oligonucleotide (lanes 5 and 6 compared to lane 4) but not with a random oligonucleotide (lanes 7 and 8). Two different specific blocking antisera were used to demonstrate the presence of MEIS1 in the upper gel shift band (compare lane 10 with lane 9 and lane 17 with lane 16). In a similar manner, specific antisera were used to demonstrate that HOXA9 (lane 11), PBX2 (lane 12), and PBX3 (lane 13) were present in this band. A combination of antisera to PBX2 plus PBX3 (lane 14) blocked almost all of the slower-migrating EMSA band. In contrast, antiserum to PBX1, which is not present in U-937 cell nuclear extracts, did not diminish the intensity of the upper band (lane 15). Since antiserum to HOXA9 blocked only a fraction of the upper gel shift band, we used specific antiserum against HOXA10 to demonstrate that this protein also contributes to the upper gel shift complex (lanes 18 and 19). Lanes 20 and 21 represent treatment of nuclear extracts (nuc. ext.) with a mixture of normal rabbit IgG and chicken IgY (r-IgG + c-IgY) and nonspecific guinea pig IgG (gp-IgG), respectively, as controls for the specific antibodies used in lanes 17 to 19. To demonstrate that the antiserum to HOXA9 blocked protein-protein interactions, in vitro-synthesized (ivt) HOXA9 and PBX1a proteins were incubated with their consensus DNA target in the absence (lane 22) or presence (lane 23) of affinity-purified antibodies to HOXA9. To demonstrate the specificity of the antibody reduction of the EMSA bands, blocking experiments were repeated using an unrelated PU.1 oligonucleotide probe previously shown to be shifted by U-937 cell extracts (43). None of the specific antibodies were able to block or supershift the EMSA bands formed with the PU.1 probe (lanes 24 to 27).
FIG. 5
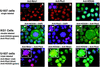
Some HOXA9 protein colocalizes with PBX2 and MEIS1 proteins to nuclear speckles in myeloid cells. (A to C) Localization of individual proteins in U-937 cells stained with affinity-purified guinea pig antibodies to MEIS1, rabbit antibodies to PBX2, and chicken antibodies to HOXA9, respectively. The insets in panels A and B show cells counterstained with DAPI to visualize nuclei; panel C shows DAPI counterstaining of cells, and the insert shows HOXA9 signal alone. Strong signals for all three proteins were detected in the nucleus, with enhanced signals within nuclear speckles. Weak staining for MEIS1 and PBX2 and moderate HOXA9 expression were observed in the cytoplasm. For each antibody, virtually all of the signal could be blocked with purified MEIS1, PBX2, or HOXA9 protein or peptide antigen, respectively (data not shown). (D to F) Double labeling with HOXA9 and PBX2 in KG1 cells. Arrows point to the green signal for HOXA9 (D), the red signal for PBX (E), or the combined yellow signal for HOXA9 plus PBX (F). (G to I) Triple labeling of U-937 cells with anti-MEIS1 (red), anti-PBX2 (blue), and anti-HOXA9 (green), showing colocalization of sets of two proteins: purple speckles resulting from the superposition of the MEIS1 and PBX signals (G), yellow speckles resulting from the superposition of the HOXA9 and MEIS1 signals (H), and aqua speckles resulting from the superposition of the HOXA9 and PBX2 signals (I). Arrows point to nuclear speckles in which all three proteins appear to be expressed.
FIG. 6
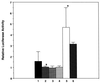
The HOXA9, MEIS1, and PBX proteins do not display transcriptional regulatory activity on a synthetic target in myeloid cells. U-937 cells were transfected with a pGL3 luciferase reporter plasmid containing a minimal SV40 promoter alone (bar 1) or with a pGL3-(PBX-HOXA9)6 plasmid containing an upstream sixfold repeat of the consensus PBX-HOXA9 binding site (bar 2). The lack of change in activity observed for the pGL3-(PBX-HOXA9)6 construct reflects the apparent inability of the endogenous HOXA9, PBX2, and MEIS1 proteins to activate or repress this reporter. Cotransfection of the pGL3-(PBX-HOXA9)6 reporter together with expression plasmids encoding the HOXA9 plus PBX2 proteins (bar 3), or HOXA9 plus PBX2 with MEIS1b proteins (bar 4), did not produce transcriptional activity. Cotransfection of the pGL3-(PBX-HOXA9)6 reporter together with HOXA9 and VP16-PBX2 expression plasmids yielded activity (bar 5), demonstrating that the reporter system was functional. Cotransfection of an expression plasmid encoding MEIS1b in addition to HOXA9 and VP16-PBX2 did not further change the observed activity (bar 6). In each assay, a cytomegalovirus-LacZ plasmid was used to normalize for transfection efficiency. ∗, P < 0.004.
Similar articles
- Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1.
Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP. Knoepfler PS, et al. Proc Natl Acad Sci U S A. 1997 Dec 23;94(26):14553-8. doi: 10.1073/pnas.94.26.14553. Proc Natl Acad Sci U S A. 1997. PMID: 9405651 Free PMC article. - Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins.
Chang CP, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML. Chang CP, et al. Mol Cell Biol. 1997 Oct;17(10):5679-87. doi: 10.1128/MCB.17.10.5679. Mol Cell Biol. 1997. PMID: 9315626 Free PMC article. - Hox cofactors in vertebrate development.
Moens CB, Selleri L. Moens CB, et al. Dev Biol. 2006 Mar 15;291(2):193-206. doi: 10.1016/j.ydbio.2005.10.032. Epub 2006 Mar 3. Dev Biol. 2006. PMID: 16515781 Review. - Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates.
Longobardi E, Penkov D, Mateos D, De Florian G, Torres M, Blasi F. Longobardi E, et al. Dev Dyn. 2014 Jan;243(1):59-75. doi: 10.1002/dvdy.24016. Epub 2013 Sep 2. Dev Dyn. 2014. PMID: 23873833 Free PMC article. Review.
Cited by
- Molecular basis of facilitated target search and sequence discrimination of TALE homeodomain transcription factor Meis1.
Choi SR, Lee J, Seo YJ, Jin HS, Ahn HB, Go Y, Kim NK, Ryu KS, Lee JH. Choi SR, et al. Nat Commun. 2024 Aug 14;15(1):6984. doi: 10.1038/s41467-024-51297-7. Nat Commun. 2024. PMID: 39143123 Free PMC article. - Novel Antennapedia and Ultrabithorax trimeric complexes with TBP and Exd regulate transcription.
Villarreal-Puente A, Altamirano-Torres C, Jiménez-Mejía G, Hernández-Bautista C, Montalvo-Méndez R, Vázquez M, Zurita M, Reséndez-Pérez D. Villarreal-Puente A, et al. Hereditas. 2024 Jul 30;161(1):25. doi: 10.1186/s41065-024-00327-x. Hereditas. 2024. PMID: 39080786 Free PMC article. - A novel fatty acid metabolism-related signature identifies features of the tumor microenvironment and predicts clinical outcome in acute myeloid leukemia.
Zhang HB, Sun ZK, Zhong FM, Yao FY, Liu J, Zhang J, Zhang N, Lin J, Li SQ, Li MY, Jiang JY, Cheng Y, Xu S, Cheng XX, Huang B, Wang XZ. Zhang HB, et al. Lipids Health Dis. 2022 Aug 25;21(1):79. doi: 10.1186/s12944-022-01687-x. Lipids Health Dis. 2022. PMID: 36002858 Free PMC article. - Trimeric complexes of Antp-TBP with TFIIEβ or Exd modulate transcriptional activity.
Jiménez-Mejía G, Montalvo-Méndez R, Hernández-Bautista C, Altamirano-Torres C, Vázquez M, Zurita M, Reséndez-Pérez D. Jiménez-Mejía G, et al. Hereditas. 2022 May 30;159(1):23. doi: 10.1186/s41065-022-00239-8. Hereditas. 2022. PMID: 35637493 Free PMC article. - MEIS1 in Hematopoiesis and Cancer. How MEIS1-PBX Interaction Can Be Used in Therapy.
Blasi F, Bruckmann C. Blasi F, et al. J Dev Biol. 2021 Oct 13;9(4):44. doi: 10.3390/jdb9040044. J Dev Biol. 2021. PMID: 34698191 Free PMC article. Review.
References
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, unit 4.1. New York, N.Y: Green Publishing Associates and Wiley Interscience; 1987.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials