A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96 - PubMed (original) (raw)
A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96
B M Fontoura et al. J Cell Biol. 1999.
Abstract
The mammalian nuclear pore complex (NPC) is comprised of approximately 50 unique proteins, collectively known as nucleoporins. Through fractionation of rat liver nuclei, we have isolated >30 potentially novel nucleoporins and have begun a systematic characterization of these proteins. Here, we present the characterization of Nup96, a novel nucleoporin with a predicted molecular mass of 96 kD. Nup96 is generated through an unusual biogenesis pathway that involves synthesis of a 186-kD precursor protein. Proteolytic cleavage of the precursor yields two nucleoporins: Nup98, a previously characterized GLFG-repeat containing nucleoporin, and Nup96. Mutational and functional analyses demonstrate that both the Nup98-Nup96 precursor and the previously characterized Nup98 (synthesized independently from an alternatively spliced mRNA) are proteolytically cleaved in vivo. This biogenesis pathway for Nup98 and Nup96 is evolutionarily conserved, as the putative Saccharomyces cerevisiae homologues, N-Nup145p and C-Nup145p, are also produced through proteolytic cleavage of a precursor protein. Using immunoelectron microscopy, Nup96 was localized to the nucleoplasmic side of the NPC, at or near the nucleoplasmic basket. The correct targeting of both Nup96 and Nup98 to the nucleoplasmic side of the NPC was found to be dependent on proteolytic cleavage, suggesting that the cleavage process may regulate NPC assembly. Finally, by biochemical fractionation, a complex containing Nup96, Nup107, and at least two Sec13- related proteins was identified, revealing that a major sub-complex of the NPC is conserved between yeast and mammals.
Figures
Figure 1
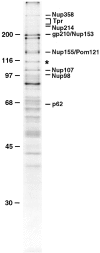
Identification of Nup96, a novel nucleoporin with an apparent molecular mass of 115 kD. Nucleoporins were purified from isolated rat liver nuclear envelopes, separated by SDS-PAGE, and stained with Coomassie brilliant blue. A previously uncharacterized protein migrating at 115 kD (indicated by an asterisk) was analyzed by peptide sequence analysis. Additional known nucleoporins are indicated. Numbers at left indicate M r markers.
Figure 5
Alternatively spliced mRNAs code for Nup98 and for a Nup98-Nup96 precursor. (A) Schematic representation of Nup98 and Nup98-Nup96 precursor mRNAs. The 5′-UTRs of both mRNAs are identical except for the first 17 nucleotides, which are absent in the Nup98-Nup96 mRNA. The Nup98 coding regions are identical, except that the Nup98-Nup96 mRNA does not contain the last 18 nucleotides coding for the COOH-terminal 6 amino acids of Nup98. The Nup98-Nup96 mRNA has an extended coding region (shown in black) that generates Nup96. Both mRNAs have different 3′-UTRs. (B) Northern blot analysis of Nup98 and Nup98-Nup96 precursor mRNA transcripts. Poly(A)+ RNA was purified from HeLa cells, separated by agarose gel electrophoresis, and transferred to nitrocellulose membranes. Membranes were probed with a Nup98-specific probe (lane 1), or a Nup96-specific probe (lane 2). Approximate size markers (in kb) are indicated on the left.
Figure 5
Alternatively spliced mRNAs code for Nup98 and for a Nup98-Nup96 precursor. (A) Schematic representation of Nup98 and Nup98-Nup96 precursor mRNAs. The 5′-UTRs of both mRNAs are identical except for the first 17 nucleotides, which are absent in the Nup98-Nup96 mRNA. The Nup98 coding regions are identical, except that the Nup98-Nup96 mRNA does not contain the last 18 nucleotides coding for the COOH-terminal 6 amino acids of Nup98. The Nup98-Nup96 mRNA has an extended coding region (shown in black) that generates Nup96. Both mRNAs have different 3′-UTRs. (B) Northern blot analysis of Nup98 and Nup98-Nup96 precursor mRNA transcripts. Poly(A)+ RNA was purified from HeLa cells, separated by agarose gel electrophoresis, and transferred to nitrocellulose membranes. Membranes were probed with a Nup98-specific probe (lane 1), or a Nup96-specific probe (lane 2). Approximate size markers (in kb) are indicated on the left.
Figure 2
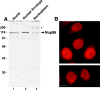
Nup96 is a component of the mammalian NPC. (A) Immunoblot analysis of fractionated rat liver nuclei. Total nuclei were fractionated into nuclear envelope and nucleoplasm, and equivalent amounts of each fraction (corresponding to 1 × 106 nuclei) were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with antibodies against Nup96. Nup96 is the major protein detected in total nuclei (lane 1), and it fractionates predominantly with the nuclear envelope fraction (lane 2). A significant amount of Nup96, however, is also detected in the nucleoplasm (lane 3). An uncharacterized, cross-reacting protein of 200 kD (indicated by the asterisk), also fractionates with the nucleoplasm. Molecular masses are indicated on the left. (B) Immunofluorescence microscopy. HeLa cells were fixed, permeabilized, and labeled with anti-Nup96 antibodies. When observed at the equatorial plane of the nucleus, a discontinuous rim was apparent, characteristic of NPC staining. Diffuse intranuclear labeling was also apparent. Lower panel shows labeling at telophase. Bar, 13 μm.
Figure 3
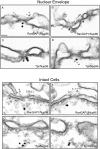
Immunolocalization of Nup96 to the nucleoplasmic face of the NPC. Isolated rat liver nuclear envelopes were probed with rabbit anti-Nup96 antibodies (A–D) and the mouse anti-RanGAP1 mAb 19C7 (A and B), or the mouse anti-Tpr mAb 5E10 (C and D). Rabbit antibodies were detected with 10 nm gold-coupled secondary antibodies, whereas mouse antibodies were detected with 5-nm gold-conjugated secondary antibodies. Envelopes were processed for thin sectioning and observed by EM. The cytoplasmic face of the nuclear envelopes (as evident from differences in the morphology of the nuclear and cytoplasmic membranes) are oriented toward the top of each micrograph. Intact HeLa cells were also probed simultaneously with rabbit anti-Nup96 antibodies (E–H) and the mouse anti-RanGAP1 mAb 19C7 (E and F), or the mouse anti-Tpr mAb 5E10 (G and H). Rabbit antibodies were detected with 10-nm gold-coupled secondary antibodies, whereas mouse antibodies were detected with 5-nm gold-conjugated secondary antibodies. The cells were processed for thin sectioning and observed by EM. The cytoplasm of the cells is oriented towards the top of each micrograph. Bar, 0.1 μm.
Figure 4
Amino acid sequence and schematic representation of the predicted Nup98-Nup96 precursor. The box indicates the NH2-terminal domain homologous to Nup98. Heavy underlines indicate amino acids identified through direct protein sequence analysis. The thin underline indicates a putative, alternative exon absent from a second class of Nup98-Nup96 precursor cDNAs. Arrow and arrowheads, between phenylalanine 863 and serine 864, indicate the proteolytic cleavage site.
Figure 7
The Nup98-Nup96 precursor is proteolytically processed in vitro and in vivo. (A) Schematic representation of the Nup98-Nup96 precursor protein constructs used in the in vitro and in vivo expression assays. Each protein contains a myc-epitope tag (shown in gray) at its NH2 terminus. (B) In vitro transcription/translation reactions. Protein constructs were transcribed and translated in rabbit reticulocyte lysates in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Reactions were performed with no DNA (lane 1), wild-type Nup98-Nup96 precursor (lane 2), mutant Nup98-Nup96 precursor (F863S/Y866R) (lane 3), Nup98 (1–863; lane 4), and Nup96 (lane 5). Molecular mass markers are indicated on the left. (C) In vivo expression of the wild-type and mutant Nup98-Nup96 precursors. HeLa cells were transfected with no DNA (lane 1), wild-type Nup98-Nup96 precursor (lane 2) and mutant Nup98-Nup96 precursor (F863S/Y866R) (lane 3). 24 h after transfection, cells were lysed in sample buffer and analyzed by immunoblot analysis using an anti-myc epitope antibody.
Figure 7
The Nup98-Nup96 precursor is proteolytically processed in vitro and in vivo. (A) Schematic representation of the Nup98-Nup96 precursor protein constructs used in the in vitro and in vivo expression assays. Each protein contains a myc-epitope tag (shown in gray) at its NH2 terminus. (B) In vitro transcription/translation reactions. Protein constructs were transcribed and translated in rabbit reticulocyte lysates in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Reactions were performed with no DNA (lane 1), wild-type Nup98-Nup96 precursor (lane 2), mutant Nup98-Nup96 precursor (F863S/Y866R) (lane 3), Nup98 (1–863; lane 4), and Nup96 (lane 5). Molecular mass markers are indicated on the left. (C) In vivo expression of the wild-type and mutant Nup98-Nup96 precursors. HeLa cells were transfected with no DNA (lane 1), wild-type Nup98-Nup96 precursor (lane 2) and mutant Nup98-Nup96 precursor (F863S/Y866R) (lane 3). 24 h after transfection, cells were lysed in sample buffer and analyzed by immunoblot analysis using an anti-myc epitope antibody.
Figure 6
Amino acid sequence alignments of the COOH-terminal portion of Nup98-Nup96 and putative homologues in S. cerevisiae (Nup145p), A. thaliana (accession number U53501), and C. elegans (accession number U50193). The box indicates the highly conserved domain preceding the cleavage site between Nup98 and Nup96. Amino acids are numbered from the presumed initiating methionine in each protein. Identical amino acids are shaded.
Figure 8
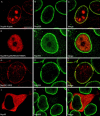
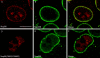
Proteolytic processing of the Nup98-Nup96 precursor is essential for targeting Nup98 and Nup96 to the NPC. HeLa cells were transiently transfected with: (A–C) the wild-type Nup98-Nup96 precursor, (D–F) the mutant Nup98-Nup96 precursor (F863S/Y866R), (G–I) the Nup98 domain (amino acids 1–863), and (J–L) Nup96 (amino acids 864–1712). Localization of the transiently expressed proteins was detected by indirect immunofluorescence and confocal microscopy using an anti-myc epitope antibody followed by a Cy3-conjugated secondary antibody (A, D, G, and J). Cells were also double labeled with an antibody specific for Nup358, which was detected using a secondary antibody conjugated to FITC (B, E, H, and K). Cy3 and FITC images were merged to identify coincident labeling of NPCs (C, F, I, and L). Bar, 10 μm.
Figure 9
Nup98 is proteolytically processed in vitro and in vivo. (A) Schematic representation of the Nup98 protein constructs used in the in vitro and in vivo expression assays. Each protein contains a myc-epitope tag (shown in gray) at its NH2 terminus. (B) In vitro transcription/translation reactions. Protein constructs were transcribed and translated in rabbit reticulocyte lysates in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Reactions were performed with no DNA (lane 1), wild-type Nup98 (lane 2), mutant Nup98 (F863S/Y866R) (lane 3), and Nup98 (1–863) (lane 4). Molecular mass markers are indicated on the left. (C) In vivo expression of the wild-type and mutant Nup98 precursors. HeLa cells were transfected wild-type Nup98 (lane 1), mutant Nup98 (F863S/Y866R) (lane 2), and with no DNA (lane 3). 24 hours after transfection, cells were lysed in sample buffer and analyzed by immunoblot analysis using an anti-myc epitope antibody.
Figure 9
Nup98 is proteolytically processed in vitro and in vivo. (A) Schematic representation of the Nup98 protein constructs used in the in vitro and in vivo expression assays. Each protein contains a myc-epitope tag (shown in gray) at its NH2 terminus. (B) In vitro transcription/translation reactions. Protein constructs were transcribed and translated in rabbit reticulocyte lysates in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Reactions were performed with no DNA (lane 1), wild-type Nup98 (lane 2), mutant Nup98 (F863S/Y866R) (lane 3), and Nup98 (1–863) (lane 4). Molecular mass markers are indicated on the left. (C) In vivo expression of the wild-type and mutant Nup98 precursors. HeLa cells were transfected wild-type Nup98 (lane 1), mutant Nup98 (F863S/Y866R) (lane 2), and with no DNA (lane 3). 24 hours after transfection, cells were lysed in sample buffer and analyzed by immunoblot analysis using an anti-myc epitope antibody.
Figure 10
Proteolytic processing of Nup98 is essential for NPC targeting. HeLa cells were transiently transfected with: (A–C) wild-type Nup98, (D–F) mutant Nup98 (F863S/Y866R). Localization of the transiently expressed proteins was detected by indirect immunofluorescence and confocal microscopy using an anti-myc epitope antibody followed by a Cy3-conjugated secondary antibody (A and D). Cells were also double labeled with an antibody specific for Nup358, which was detected using a secondary antibody conjugated to FITC (B and E). Cy3 and FITC images were merged to identify coincident labeling of NPCs (C and F). Bar, 10 μm.
Figure 11
Identification of a complex containing Nup96, Nup107, and Sec13. Rat liver nuclei were fractionated as described in Materials and Methods. A fraction containing soluble NPCs was loaded onto a 10–40% sucrose gradient and centrifuged. Gradient fractions were collected and analyzed by SDS-PAGE followed by silver stain analysis (top), or immunoblot analysis with antibodies specific for Nup96 and Nup107 (bottom). Fractions, numbered from the top of the gradient, are indicated. Asterisks between lanes 5 and 6 (top) indicate proteins that sediment as a complex. Some of the proteins comprising this complex are indicated on the right.
Similar articles
- The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex.
Radu A, Moore MS, Blobel G. Radu A, et al. Cell. 1995 Apr 21;81(2):215-22. doi: 10.1016/0092-8674(95)90331-3. Cell. 1995. PMID: 7736573 - Autoproteolysis in nucleoporin biogenesis.
Rosenblum JS, Blobel G. Rosenblum JS, et al. Proc Natl Acad Sci U S A. 1999 Sep 28;96(20):11370-5. doi: 10.1073/pnas.96.20.11370. Proc Natl Acad Sci U S A. 1999. PMID: 10500183 Free PMC article. - Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins.
Teixeira MT, Siniossoglou S, Podtelejnikov S, Bénichou JC, Mann M, Dujon B, Hurt E, Fabre E. Teixeira MT, et al. EMBO J. 1997 Aug 15;16(16):5086-97. doi: 10.1093/emboj/16.16.5086. EMBO J. 1997. PMID: 9305650 Free PMC article. - The nuclear pore complex.
Davis LI. Davis LI. Annu Rev Biochem. 1995;64:865-96. doi: 10.1146/annurev.bi.64.070195.004245. Annu Rev Biochem. 1995. PMID: 7574503 Review. - Interdependence between Nuclear Pore Gatekeepers and Genome Caretakers: Cues from Genome Instability Syndromes.
Larizza L, Colombo EA. Larizza L, et al. Int J Mol Sci. 2024 Aug 29;25(17):9387. doi: 10.3390/ijms25179387. Int J Mol Sci. 2024. PMID: 39273335 Free PMC article. Review.
Cited by
- The death domain-containing protein Unc5CL is a novel MyD88-independent activator of the pro-inflammatory IRAK signaling cascade.
Heinz LX, Rebsamen M, Rossi DC, Staehli F, Schroder K, Quadroni M, Gross O, Schneider P, Tschopp J. Heinz LX, et al. Cell Death Differ. 2012 Apr;19(4):722-31. doi: 10.1038/cdd.2011.147. Epub 2011 Dec 9. Cell Death Differ. 2012. PMID: 22158417 Free PMC article. - "Off-pore" nucleoporins relocalize heterochromatic breaks through phase separation.
Merigliano C, Ryu T, See CD, Caridi CP, Li X, Butova NL, Reynolds TW, Deng C, Chenoweth DM, Capelson M, Chiolo I. Merigliano C, et al. bioRxiv [Preprint]. 2024 Jul 18:2023.12.07.570729. doi: 10.1101/2023.12.07.570729. bioRxiv. 2024. PMID: 39071440 Free PMC article. Preprint. - Advances in the understanding of nuclear pore complexes in human diseases.
Li Y, Zhu J, Zhai F, Kong L, Li H, Jin X. Li Y, et al. J Cancer Res Clin Oncol. 2024 Jul 30;150(7):374. doi: 10.1007/s00432-024-05881-5. J Cancer Res Clin Oncol. 2024. PMID: 39080077 Free PMC article. Review. - SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling.
Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, Cupic A, Makio T, Mei M, Moreno E, Danziger O, White KM, Rathnasinghe R, Uccellini M, Gao S, Aydillo T, Mena I, Yin X, Martin-Sancho L, Krogan NJ, Chanda SK, Schotsaert M, Wozniak RW, Ren Y, Rosenberg BR, Fontoura BMA, García-Sastre A. Miorin L, et al. Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):28344-28354. doi: 10.1073/pnas.2016650117. Epub 2020 Oct 23. Proc Natl Acad Sci U S A. 2020. PMID: 33097660 Free PMC article. - Human nucleoporins promote HIV-1 docking at the nuclear pore, nuclear import and integration.
Di Nunzio F, Danckaert A, Fricke T, Perez P, Fernandez J, Perret E, Roux P, Shorte S, Charneau P, Diaz-Griffero F, Arhel NJ. Di Nunzio F, et al. PLoS One. 2012;7(9):e46037. doi: 10.1371/journal.pone.0046037. Epub 2012 Sep 25. PLoS One. 2012. PMID: 23049930 Free PMC article.
References
- Bastos, R., N. Pante, and B. Burke. 1995. Nuclear pore complex proteins. Int. Rev. Cytol. 162B:257–302. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous