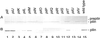Twelve pil genes are required for biogenesis of the R64 thin pilus - PubMed (original) (raw)
Twelve pil genes are required for biogenesis of the R64 thin pilus
T Yoshida et al. J Bacteriol. 1999 Apr.
Abstract
The IncI1 plasmid R64 produces two kinds of sex pili: a thin pilus and a thick pilus. The thin pilus, which belongs to the type IV family, is required only for liquid matings. Fourteen genes, pilI to -V, were found in the DNA region responsible for the biogenesis of the R64 thin pilus (S.-R. Kim and T. Komano, J. Bacteriol. 179:3594-3603, 1997). In this study, we introduced frameshift mutations into each of the 14 pil genes to test their requirement for R64 thin pilus biogenesis. From the analyses of extracellular secretion of thin pili and transfer frequency in liquid matings, we found that 12 genes, pilK to -V, are required for the formation of the thin pilus. Complementation experiments excluded the possible polar effects of each mutation on the expression of downstream genes. Two genes, traBC, were previously shown to be required for the expression of the pil genes. In addition, the rci gene is responsible for modulating the structure and function of the R64 thin pilus via the DNA rearrangement of the shufflon. Altogether, 15 genes, traBC, pilK through pilV, and rci, are essential for R64 thin pilus formation and function.
Figures
FIG. 1
Gene organization of the traA to -D and pilI to -V regions of pKK641-A′. The top horizontal line represents a restriction map. B, _Bgl_II; C, _Cla_I; E, _Eco_RI; H, _Hin_dIII; Hp, _Hpa_I; P, _Pst_I; S, _Sma_I; V, _Pvu_II. Locations of insertion or deletion mutations and their transfer capacity are indicated (open circles, transfer proficient; solid circles, transfer deficient). Below the map, open reading frames are represented by arrows. tra, transfer; pil, formation of thin pilus; Shf, shufflon; rci, shufflon-specific recombinase. The solid lines on the bottom indicate DNA portions present in complementing plasmids.
FIG. 2
(A) Effects of various pil mutations on the expression and processing of the pilS product. Whole extracts of E. coli cells harboring pKK641A′ with the indicated pil mutations were separated by SDS-PAGE and subjected to Western blot analysis with antipilin antiserum. (B) Effects of pil mutations on the formation of thin pili. A thin pilus fraction was prepared from the culture media of E. coli cells harboring pKK641A′ with the indicated pil mutations. Proteins were separated by SDS-PAGE and subjected to Western blot analysis with antipilin antiserum. The positions of prepilin (22 kDa) and pilin (19 kDa) are indicated on the right.
Similar articles
- The plasmid R64 thin pilus identified as a type IV pilus.
Kim SR, Komano T. Kim SR, et al. J Bacteriol. 1997 Jun;179(11):3594-603. doi: 10.1128/jb.179.11.3594-3603.1997. J Bacteriol. 1997. PMID: 9171405 Free PMC article. - Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili.
Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T. Yoshida T, et al. J Bacteriol. 1998 Jun;180(11):2842-8. doi: 10.1128/JB.180.11.2842-2848.1998. J Bacteriol. 1998. PMID: 9603870 Free PMC article. - Genes required for plasmid R64 thin-pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes.
Sakai D, Komano T. Sakai D, et al. J Bacteriol. 2002 Jan;184(2):444-51. doi: 10.1128/JB.184.2.444-451.2002. J Bacteriol. 2002. PMID: 11751821 Free PMC article. - Mating variation by DNA inversions of shufflon in plasmid R64.
Komano T, Kim SR, Yoshida T. Komano T, et al. Adv Biophys. 1995;31:181-93. doi: 10.1016/0065-227x(95)99391-2. Adv Biophys. 1995. PMID: 7625273 Review. - Structure and function of the shufflon in plasmid R64.
Gyohda A, Furuya N, Ishiwa A, Zhu S, Komano T. Gyohda A, et al. Adv Biophys. 2004;38:183-213. Adv Biophys. 2004. PMID: 15493334 Review.
Cited by
- Salmonella enterica serovar typhi uses type IVB pili to enter human intestinal epithelial cells.
Zhang XL, Tsui IS, Yip CM, Fung AW, Wong DK, Dai X, Yang Y, Hackett J, Morris C. Zhang XL, et al. Infect Immun. 2000 Jun;68(6):3067-73. doi: 10.1128/IAI.68.6.3067-3073.2000. Infect Immun. 2000. PMID: 10816445 Free PMC article. - Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives.
Berry JL, Pelicic V. Berry JL, et al. FEMS Microbiol Rev. 2015 Jan;39(1):134-54. doi: 10.1093/femsre/fuu001. Epub 2014 Dec 4. FEMS Microbiol Rev. 2015. PMID: 25793961 Free PMC article. Review. - The Type IV Pilus of Plasmid TP114 Displays Adhesins Conferring Conjugation Specificity and Is Important for DNA Transfer in the Mouse Gut Microbiota.
Allard N, Neil K, Grenier F, Rodrigue S. Allard N, et al. Microbiol Spectr. 2022 Apr 27;10(2):e0230321. doi: 10.1128/spectrum.02303-21. Epub 2022 Mar 16. Microbiol Spectr. 2022. PMID: 35293798 Free PMC article. - Incompatibility Group I1 (IncI1) Plasmids: Their Genetics, Biology, and Public Health Relevance.
Foley SL, Kaldhone PR, Ricke SC, Han J. Foley SL, et al. Microbiol Mol Biol Rev. 2021 Apr 28;85(2):e00031-20. doi: 10.1128/MMBR.00031-20. Print 2021 May 19. Microbiol Mol Biol Rev. 2021. PMID: 33910982 Free PMC article. Review. - Structural and evolutionary analyses show unique stabilization strategies in the type IV pili of Clostridium difficile.
Piepenbrink KH, Maldarelli GA, Martinez de la Peña CF, Dingle TC, Mulvey GL, Lee A, von Rosenvinge E, Armstrong GD, Donnenberg MS, Sundberg EJ. Piepenbrink KH, et al. Structure. 2015 Feb 3;23(2):385-96. doi: 10.1016/j.str.2014.11.018. Epub 2015 Jan 15. Structure. 2015. PMID: 25599642 Free PMC article.
References
- Alm R A, Hallinan J P, Watson A A, Mattick J S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. - PubMed
- Bradley D E. Derepressed plasmids of incompatibility group I1 determine two different morphological forms of pilus. Plasmid. 1983;9:331–334. - PubMed
- Bradley D E. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K, and Z. J Gen Microbiol. 1984;130:1489–1502. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources