Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand - PubMed (original) (raw)
. 1999 Mar 30;96(7):3540-5.
doi: 10.1073/pnas.96.7.3540.
D L Lacey, C R Dunstan, I Solovyev, A Colombero, E Timms, H L Tan, G Elliott, M J Kelley, I Sarosi, L Wang, X Z Xia, R Elliott, L Chiu, T Black, S Scully, C Capparelli, S Morony, G Shimamoto, M B Bass, W J Boyle
Affiliations
- PMID: 10097072
- PMCID: PMC22329
- DOI: 10.1073/pnas.96.7.3540
Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand
H Hsu et al. Proc Natl Acad Sci U S A. 1999.
Abstract
A receptor that mediates osteoprotegerin ligand (OPGL)-induced osteoclast differentiation and activation has been identified via genomic analysis of a primary osteoclast precursor cell cDNA library and is identical to the tumor necrosis factor receptor (TNFR) family member RANK. The RANK mRNA was highly expressed by isolated bone marrow-derived osteoclast progenitors and by mature osteoclasts in vivo. Recombinant OPGL binds specifically to RANK expressed by transfected cell lines and purified osteoclast progenitors. Transgenic mice expressing a soluble RANK-Fc fusion protein have severe osteopetrosis because of a reduction in osteoclasts, similar to OPG transgenic mice. Recombinant RANK-Fc binds with high affinity to OPGL in vitro and blocks osteoclast differentiation and activation in vitro and in vivo. Furthermore, polyclonal Ab against the RANK extracellular domain promotes osteoclastogenesis in bone marrow cultures suggesting that RANK activation mediates the effects of OPGL on the osteoclast pathway. These data indicate that OPGL-induced osteoclastogenesis is directly mediated through RANK on osteoclast precursor cells.
Figures
Figure 1
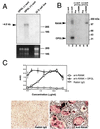
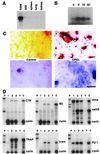
RANK mediates OPGL-induced osteoclastogenesis. (A) Expression of RANK in osteoclast precursor cells. A RANK-specific probe was used in the Northern blot analysis of 4 μg of total RNA prepared from the negative OPGL-binding population, positive OPGL-binding population, or 32-D cells. (B) Cross-linking of OPGL–RANK complexes on the surface of osteoclast precursors. OPGL positive and negative binding cells were prepared by FACS, incubated with 125I-labeled OPGL, and then subjected to cross-linking with 1 mM disuccinimidyl tartrate. One-tenth of the cell lysates were directly fractionated by SDS/PAGE (lanes 1 and 2). The remainder of the cell lysates were immunoprecipitated with either pre-immune serum (lanes 3 and 5) or anti-RANK Ab (lanes 4 and 6). (C) Anti-RANK Abs stimulate osteoclastogenesis. Mouse bone marrow cells were cultured with CSF-1 (30 ng/ml) with and without 5 ng/ml murine OPGL(158–316) in the presence of various concentrations of either anti-RANK Ab or rabbit IgG. Osteoclastogenesis was quantified by TRAP solution assay. A405 represents absorbance at 405 nm and is presented as the mean ± SD (n = 3). TRAP cytochemical staining was shown at the bottom. (Bar = 200 μm.)
Figure 2
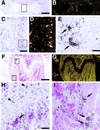
Localization of RANK mRNA expression in developing and adult bone by in situ hybridization. Sections of embryonic and adult long bones were hybridized with 33P-labeled antisense riboprobes to RANK, processed and stained with hematoxylin/eosin. Panels A–E are from fetal (E15.5) femur, with B and D being darkfield images of A and C, respectively. Panels F–I are from adult (6-week) femur, with G being a darkfield image of F. RANK is expressed by multinucleated cells attached to matrix, consistent with osteoclasts in areas of active bone modeling in the fetus and adult. Hypertrophic chondrocytes in the adult growth plate also appear to express RANK transcripts. Arrows mark the position of osteoclasts, whereas arrowheads mark the position of hypertrophic chondrocytes. [Bars = 400 μm (A and F), 100 μm (C), and 25 μm (E and H).]
Figure 3
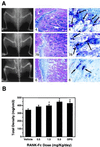
Soluble RANK-Fc increases bone density. (A) Increased bone density in RANK-Fc transgenic mice. A high RANK-Fc expressing mouse (animal 6), medium expressing mouse (animal 78), and control littermate (animal 4) were subjected to radiographic analysis (Left) and histologic analysis (Middle and Right). Increased radiodensity of the long bones, pelvic bones, and the vertebrae were detected in both the high and low expressors (Left) with no changes in bone shape. The femoral diaphyses of the RANK-Fc transgenic mice contained nonresorbed bone and cartilage with the highest expressor (animal 6) having the greatest accumulation (Middle). TRAP histochemical staining of the bones showed decreased osteoclast numbers in the RANK-Fc expressors (Right). (B) PQCT analysis of mice treated with RANK-Fc recombinant protein. Male BDF1 mice aged 3 weeks received the indicated doses of RANK-Fc fusion protein by single daily subcutaneous injection for 4 days. The bone density in the proximal tibial metaphysis was measured by peripheral quantitative computerized tomography. Total density was presented as the mean ± SD (n = 4). Treatment with RANK-Fc induced a dose-dependent increase of total bone density (P < 0.001).
Figure 4
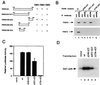
RANK interactions with TRAF proteins and activation of NF-κB and JNK pathways. (A) Mapping of TRAF-binding domains of RANK. Expression vectors encoding full-length or deletion mutants of RANK intracellular domain fused to the GAL4 DNA-binding domain were cotransformed into HF7C yeast strain with vectors expressing the GAL4 activation domain fused with TRAF2, TRAF5, or TRAF6 as described (21). +, Growth after 1 week on the selection plates. (B) Coimmunoprecipitation of RANK and TRAF proteins. After 24 hr of transfection, 293 cell lysates were immunoprecipitated with pre-immune serum or Ab against RANK extracellular domain. Coprecipitating TRAF2 or TRAF6 were detected by immunoblot analysis with goat anti-TRAF2 or anti-TRAF6 Ab (Santa Cruz Biotechnology). (C) Activation of NF-κB in RANK transfected 293 cells. Luciferase activities were measured after 24 hr and normalized on the basis of β-galactosidase expression levels. The values (means ± SD) represent luciferase activities relative to vector transfection for representative experiments performed in duplicate. (D) Activation of JNK in RANK-transfected 293 cells. After 24-hr transfection, 293 cell lysates were immunoprecipitated with monoclonal anti-HA epitope Ab. Immunoprecipitates were assayed for kinase activity by using 2 μg GST-JUN (Stratagene) as substrate, and resolved by SDS/PAGE.
Figure 5
OPGL and anti-RANK agonistic Abs stimulate osteoclast differentiation of the murine myeloid RAW 264.7 cell line. (A) Northern blots of RNA from various sources hybridized with an RANK probe. (B) Approximately 106 RAW cells were exposed to 100 ng/ml OPGL for the indicated length of time. The cell lysates were immunoprecipitated with monoclonal anti-JNK Ab. Immunoprecipitates were assayed for kinase activity by using GST-JUN as substrate and resolved by SDS/PAGE. (C) RAW cells were plated on bone slices and treated for 7 days with OPGL. The Upper and Lower panels show the results of TRAP cytochemistry and toluidine blue staining, respectively. Note that OPGL induces the formation of TRAP positive cells (Upper) that form resorption lacunae (Lower). (Bars = 50 μm.) (D) Total cell RNA was extracted from RAW cells treated with media alone (lane 1) or media supplemented with either 100 ng/ml OPGL (lane 2) or 1,750 ng/ml anti-RANK antibodies (lane 3). RNA from OPGL plus CSF-1-treated bone marrow cultures (lane C) served as the positive control. RNase protection assays were performed by using probes for the calcitonin receptor (CTR), β3 integrin (B3), c-fms, cathepsin k (cathK), TRAP, c-src, PU.1, and cyclophilin (cyclo). Lanes P and Y represent the undigested probe and yeast hybridization controls, respectively.
Similar articles
- The ligand for osteoprotegerin (OPGL) directly activates mature osteoclasts.
Burgess TL, Qian Y, Kaufman S, Ring BD, Van G, Capparelli C, Kelley M, Hsu H, Boyle WJ, Dunstan CR, Hu S, Lacey DL. Burgess TL, et al. J Cell Biol. 1999 May 3;145(3):527-38. doi: 10.1083/jcb.145.3.527. J Cell Biol. 1999. PMID: 10225954 Free PMC article. - Osteoprotegerin and osteoprotegerin ligand effects on osteoclast formation from human peripheral blood mononuclear cell precursors.
Shalhoub V, Faust J, Boyle WJ, Dunstan CR, Kelley M, Kaufman S, Scully S, Van G, Lacey DL. Shalhoub V, et al. J Cell Biochem. 1999 Feb 1;72(2):251-61. J Cell Biochem. 1999. PMID: 10022507 - A new member of tumor necrosis factor ligand family, ODF/OPGL/TRANCE/RANKL, regulates osteoclast differentiation and function.
Takahashi N, Udagawa N, Suda T. Takahashi N, et al. Biochem Biophys Res Commun. 1999 Mar 24;256(3):449-55. doi: 10.1006/bbrc.1999.0252. Biochem Biophys Res Commun. 1999. PMID: 10080918 Review. - Osteoprotegerin: a physiological and pharmacological inhibitor of bone resorption.
Kostenuik PJ, Shalhoub V. Kostenuik PJ, et al. Curr Pharm Des. 2001 May;7(8):613-35. doi: 10.2174/1381612013397807. Curr Pharm Des. 2001. PMID: 11375772 Review.
Cited by
- Improved osteogenesis in rat femur segmental defects treated with human allograft and zinc adjuvants.
Kanjilal D, Grieg C, Culbertson MD, Lin SS, Vives M, Benevenia J, O'Connor JP. Kanjilal D, et al. Exp Biol Med (Maywood). 2021 Aug;246(16):1857-1868. doi: 10.1177/15353702211019008. Epub 2021 May 26. Exp Biol Med (Maywood). 2021. PMID: 34038225 Free PMC article. - Assessing cancer pain.
Dalal S, Bruera E. Dalal S, et al. Curr Pain Headache Rep. 2012 Aug;16(4):314-24. doi: 10.1007/s11916-012-0274-y. Curr Pain Headache Rep. 2012. PMID: 22585314 Review. - Monocytes/Macrophages Upregulate the Hyaluronidase HYAL1 and Adapt Its Subcellular Trafficking to Promote Extracellular Residency upon Differentiation into Osteoclasts.
Puissant E, Boonen M. Puissant E, et al. PLoS One. 2016 Oct 18;11(10):e0165004. doi: 10.1371/journal.pone.0165004. eCollection 2016. PLoS One. 2016. PMID: 27755597 Free PMC article. - Painful boney metastases.
Smith HS, Mohsin I. Smith HS, et al. Korean J Pain. 2013 Jul;26(3):223-41. doi: 10.3344/kjp.2013.26.3.223. Epub 2013 Jul 1. Korean J Pain. 2013. PMID: 23861996 Free PMC article. - Inhibition of osteoclast generation: a novel function of the bone morphogenetic protein 7/osteogenic protein 1.
Maurer T, Zimmermann G, Maurer S, Stegmaier S, Wagner C, Hänsch GM. Maurer T, et al. Mediators Inflamm. 2012;2012:171209. doi: 10.1155/2012/171209. Epub 2012 Oct 24. Mediators Inflamm. 2012. PMID: 23132958 Free PMC article.
References
- Suda T, Takahashi N, Martin T J. Endocr Rev. 1992;13:66–80. - PubMed
- Roodman G D. Endocr Rev. 1996;17:308–332. - PubMed
- Yasuda H, Shima N, Nakagawa N, Mochizuki S I, Yano K, Fujise N, Sato Y, Goo M, Yamaguchi K, Kuriyama M, et al. Endocrinology. 1998;139:1329–1337. - PubMed
- Simonet W S, Lacey D L, Kelley M, Chang M S, Luthy R, Nguyen H, Wooden S, Bennett L, Dunstan C, Boone T, et al. Cell. 1997;89:309–319. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases