Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta - PubMed (original) (raw)
Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta
L A Paige et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Estrogen receptor (ER) modulators produce distinct tissue-specific biological effects, but within the confines of the established models of ER action it is difficult to understand why. Previous studies have suggested that there might be a relationship between ER structure and activity. Different ER modulators may induce conformational changes in the receptor that result in a specific biological activity. To investigate the possibility of modulator-specific conformational changes, we have applied affinity selection of peptides to identify binding surfaces that are exposed on the apo-ERs alpha and beta and on each receptor complexed with estradiol or 4-OH tamoxifen. These peptides are sensitive probes of receptor conformation. We show here that ER ligands, known to produce distinct biological effects, induce distinct conformational changes in the receptors, providing a strong correlation between ER conformation and biological activity. Furthermore, the ability of some of the peptides to discriminate between different ER alpha and ER beta ligand complexes suggests that the biological effects of ER agonists and antagonists acting through these receptors are likely to be different.
Figures
Figure 1
(A) Sequences of LXXLL motif containing peptides that were affinity selected on ER α in the presence of estradiol. (B) Sequences of LXXLL motifs found in the nuclear receptor coactivating proteins human SRC1a (steroid receptor coactivator 1a), mouse cAMP-responsive element binding protein (CREB)-binding protein (CBP), and human RIP140.
Figure 2
Phage ELISA. A biotinylated vitellogenin ERE was immobilized on 96-well plates precoated with streptavidin. The ER was then immobilized on the ERE and incubated for 5 min in the presence of modulator before the addition of phage. Assays were conducted as described in Materials and Methods. HRP, horseradish peroxidase.
Figure 3
Analysis of the binding specificity of the conformational probes was conducted by phage ELISA as described in Materials and Methods. Estradiol and 4-OH tamoxifen concentrations were 1 μM. The probes α/β I-α/β V are shown only for ER α. The binding patterns of these probes on ER β were similar. Sequences of the probes are as follows: α/β I, SSNHQSSRLIELLSR; α/β II, SAPRATISHYLMGG; α/β III, SSWDMHQFFWEGVSR; α/β IV, SRLPPSVFSMCGSEVCLSR; α/β V, SSPGSREWFKDMLSR; α I, SSEYCFYWDSAHCSR; α II, SSLTSRDFGSWYASR; α III, SRTWESPLGTWEWSR; β I, SREWEDGFGGRWLSR; β II, SSLDLSQFPMTASFLRESR; β III, SSEACVGRWMLCEQLGVSR.
Figure 4
Fingerprint analysis of ER modulators on (A) ER α and (B) ER β. Immobilized ER was incubated with estradiol (1 μM), estriol (1 μM), Premarin (10 μM), 4-OH tamoxifen (1 μM), nafoxidine (10 μM), clomiphene (10 μM), raloxifene (1 μM), ICI 182,780 (1 μM), 16α-OH estrone (10 μM), DES (1 μM), or progesterone (1 μM). Phage ELISAs were conducted as described in Materials and Methods.
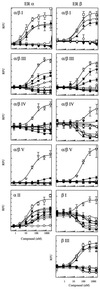
Figure 5
Comparison of the binding of the peptide probes to ER α or ER β in the presence of modulators using TRF. Assays were conducted as described in Materials and Methods. (○), buffer, aporeceptor; (□) 17β estradiol;(▵) estriol; (▴) DES; (▿) 4-OH tamoxifen; (▾) raloxifene; (⋄)nafoxidine; (♦) clomiphene; (★) ICI 182,780. RFU, relative fluorescence units.
Similar articles
- A naturally occurring estrogen receptor mutation results in increased estrogenicity of a tamoxifen analog.
Catherino WH, Wolf DM, Jordan VC. Catherino WH, et al. Mol Endocrinol. 1995 Aug;9(8):1053-63. doi: 10.1210/mend.9.8.7476979. Mol Endocrinol. 1995. PMID: 7476979 - Yeast two-hybrid system demonstrates that estrogen receptor dimerization is ligand-dependent in vivo.
Wang H, Peters GA, Zeng X, Tang M, Ip W, Khan SA. Wang H, et al. J Biol Chem. 1995 Oct 6;270(40):23322-9. doi: 10.1074/jbc.270.40.23322. J Biol Chem. 1995. PMID: 7559488 - Peptide antagonists of the human estrogen receptor.
Norris JD, Paige LA, Christensen DJ, Chang CY, Huacani MR, Fan D, Hamilton PT, Fowlkes DM, McDonnell DP. Norris JD, et al. Science. 1999 Jul 30;285(5428):744-6. doi: 10.1126/science.285.5428.744. Science. 1999. PMID: 10426998 - Molecular mechanisms of estrogen action: selective ligands and receptor pharmacology.
Katzenellenbogen BS, Choi I, Delage-Mourroux R, Ediger TR, Martini PG, Montano M, Sun J, Weis K, Katzenellenbogen JA. Katzenellenbogen BS, et al. J Steroid Biochem Mol Biol. 2000 Nov 30;74(5):279-85. doi: 10.1016/s0960-0760(00)00104-7. J Steroid Biochem Mol Biol. 2000. PMID: 11162936 Review. - Molecular conformation, receptor binding, and hormone action of natural and synthetic estrogens and antiestrogens.
Duax WL, Griffin JF, Weeks CM, Korach KS. Duax WL, et al. Environ Health Perspect. 1985 Sep;61:111-21. doi: 10.1289/ehp.8561111. Environ Health Perspect. 1985. PMID: 3905370 Free PMC article. Review.
Cited by
- Detrimental Effects of Bisphenol Compounds on Physiology and Reproduction in Fish: A Literature Review.
Faheem M, Bhandari RK. Faheem M, et al. Environ Toxicol Pharmacol. 2021 Jan;81:103497. doi: 10.1016/j.etap.2020.103497. Epub 2020 Sep 17. Environ Toxicol Pharmacol. 2021. PMID: 32950715 Free PMC article. Review. - Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor.
Koide A, Abbatiello S, Rothgery L, Koide S. Koide A, et al. Proc Natl Acad Sci U S A. 2002 Feb 5;99(3):1253-8. doi: 10.1073/pnas.032665299. Epub 2002 Jan 29. Proc Natl Acad Sci U S A. 2002. PMID: 11818562 Free PMC article. - Metalloprotein switches that display chemical-dependent electron transfer in cells.
Atkinson JT, Campbell IJ, Thomas EE, Bonitatibus SC, Elliott SJ, Bennett GN, Silberg JJ. Atkinson JT, et al. Nat Chem Biol. 2019 Feb;15(2):189-195. doi: 10.1038/s41589-018-0192-3. Epub 2018 Dec 17. Nat Chem Biol. 2019. PMID: 30559426 Free PMC article. - Estrogen stimuli promote osteoblastic differentiation via the subtilisin-like proprotein convertase PACE4 in MC3T3-E1 cells.
Kim H, Tabata A, Tomoyasu T, Ueno T, Uchiyama S, Yuasa K, Tsuji A, Nagamune H. Kim H, et al. J Bone Miner Metab. 2015 Jan;33(1):30-9. doi: 10.1007/s00774-014-0567-9. Epub 2014 Feb 21. J Bone Miner Metab. 2015. PMID: 24557631 - Steroid receptor coactivators 1, 2, and 3: critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy.
Johnson AB, O'Malley BW. Johnson AB, et al. Mol Cell Endocrinol. 2012 Jan 30;348(2):430-9. doi: 10.1016/j.mce.2011.04.021. Epub 2011 Jun 1. Mol Cell Endocrinol. 2012. PMID: 21664237 Free PMC article. Review.
References
- Evans R M, Hollenberg S M. Cell. 1988;52:1–3. - PubMed
- Smith D F, Toft D O. Mol Endocrinol. 1993;7:4–11. - PubMed
- Onate S A, Tsai S, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. - PubMed
- Norris J D, Fan D, Stallcup M R, McDonnell D P. J Biol Chem. 1998;273:6679–6688. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources