A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells - PubMed (original) (raw)
A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells
S Rajagopalan et al. J Exp Med. 1999.
Erratum in
- J Exp Med 2000 Jun 5;191(11):following 2027
Abstract
Human natural killer (NK) cells express several killer cell immunoglobulin (Ig)-like receptors (KIRs) that inhibit their cytotoxicity upon recognition of human histocompatibility leukocyte antigen (HLA) class I molecules on target cells. Additional members of the KIR family, including some that deliver activation signals, have unknown ligand specificity and function. One such KIR, denoted KIR2DL4, is structurally divergent from other KIRs in the configuration of its two extracellular Ig domains and of its transmembrane and cytoplasmic domains. Here we show that recombinant soluble KIR2DL4 binds to cells expressing HLA-G but not to cells expressing other HLA class I molecules. Unlike other HLA class I-specific KIRs, which are clonally distributed on NK cells, KIR2DL4 is expressed at the surface of all NK cells. Furthermore, functional transfer of KIR2DL4 into the cell line NK-92 resulted in inhibition of lysis of target cells that express HLA-G, but not target cells that express other class I molecules including HLA-E. Therefore, given that HLA-G expression is restricted to fetal trophoblast cells, KIR2DL4 may provide important signals to maternal NK decidual cells that interact with trophoblast cells at the maternal-fetal interface during pregnancy.
Figures
Figure 1
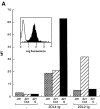
Binding of soluble KIR2DL4–Ig to 221–G cells. (A) 721.221 (221) and 721.221 transfectants expressing HLA-Cw*0304 (221-Cw3) or HLA-G (221-G) were incubated with no fusion protein, 25 μg/ml of 2DL2–Ig, or 25 μg/ml of 2DL4–Ig. The bound fusion proteins were detected by flow cytometry after reaction with FITC-conjugated goat anti– human Fc specific antibodies. Data are expressed as median fluorescence intensity (MFI). Inset, histogram profile (filled) of ungated 221-G cells stained with 2DL4–Ig. Open histogram is control staining with secondary antibodies alone. (B) Binding of KIR2DL4–Ig to 721.221 transfectants was detected as described in A. Cell surface expression of HLA class I on the different cells, as detected by staining with the mAb DX17, was as follows (MFI in parenthesis): 221 (21); 221–B7 (1,731); 221–Cw3 (588); and 221–G (1,540).
Figure 1
Binding of soluble KIR2DL4–Ig to 221–G cells. (A) 721.221 (221) and 721.221 transfectants expressing HLA-Cw*0304 (221-Cw3) or HLA-G (221-G) were incubated with no fusion protein, 25 μg/ml of 2DL2–Ig, or 25 μg/ml of 2DL4–Ig. The bound fusion proteins were detected by flow cytometry after reaction with FITC-conjugated goat anti– human Fc specific antibodies. Data are expressed as median fluorescence intensity (MFI). Inset, histogram profile (filled) of ungated 221-G cells stained with 2DL4–Ig. Open histogram is control staining with secondary antibodies alone. (B) Binding of KIR2DL4–Ig to 721.221 transfectants was detected as described in A. Cell surface expression of HLA class I on the different cells, as detected by staining with the mAb DX17, was as follows (MFI in parenthesis): 221 (21); 221–B7 (1,731); 221–Cw3 (588); and 221–G (1,540).
Figure 2
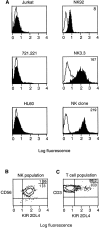
Cell surface expression of KIR2DL4 on NK cells. (A) Flow cytometry profiles of the T leukemia cell line Jurkat, the B lymphoblastoid cell line 721.221, the monocytic cell line HL60 and the NK cell lines NK-92 and NK3.3 as well as a CD3−CD56+CD94− NK clone. Cells were incubated with anti-2DL4 antiserum followed by FITC anti–rabbit IgG. (B) Flow cytometry analysis of CD3−CD56+ NK cells isolated from the peripheral blood of a normal donor and double stained with PE-conjugated CD56 and rabbit anti-2DL4 antiserum followed by FITC-conjugated goat anti–rabbit IgG. The percentages of total cells in each quadrant are listed. (C) Flow cytometry analysis of a primary T cell culture from a normal donor gated on CD3+ cells and double stained for PE-conjugated CD3 and rabbit anti-2DL4 followed by FITC-conjugated goat anti–rabbit IgG. The percentages of total cells in each quadrant are listed.
Figure 3
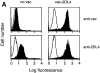
Functional transfer of KIR2DL4 in NK-92 cells results in HLA-G–mediated inhibition. (A) Flow cytometry analysis of cell surface KIR2DL4 in NK-92 cells either uninfected, or infected with vac-2DL4 (15 PFU/cell). The cells were stained with either anti-2DL4 rabbit antiserum followed by FITC goat anti–rabbit IgG or with the mAb VV1-1G10 specific for a vaccinia cell surface protein. The histograms show log fluorescence of ungated cells. (B) Aliquots of uninfected or vac-2DL4–infected cells in A were tested for their ability to lyse 721.221 cells and 721.221 cells transfected with HLA-Cw*0401 and HLA-G, in a 4-h 51Cr-release assay. Lysis is shown for an E/T ratio of 5. Similar data was obtained at other E/T ratios and in five independent experiments. (C) Aliquots of NK-92 cells infected with vac-2DL4 were tested for their ability to lyse 721.221 (squares) or 221-G cells (triangles) in the presence of 5 μg/ml of the anti-class I mAb DX17 (open symbols) or control IgG1 (filled symbols). Similar results were obtained in two independent experiments.
Figure 3
Functional transfer of KIR2DL4 in NK-92 cells results in HLA-G–mediated inhibition. (A) Flow cytometry analysis of cell surface KIR2DL4 in NK-92 cells either uninfected, or infected with vac-2DL4 (15 PFU/cell). The cells were stained with either anti-2DL4 rabbit antiserum followed by FITC goat anti–rabbit IgG or with the mAb VV1-1G10 specific for a vaccinia cell surface protein. The histograms show log fluorescence of ungated cells. (B) Aliquots of uninfected or vac-2DL4–infected cells in A were tested for their ability to lyse 721.221 cells and 721.221 cells transfected with HLA-Cw*0401 and HLA-G, in a 4-h 51Cr-release assay. Lysis is shown for an E/T ratio of 5. Similar data was obtained at other E/T ratios and in five independent experiments. (C) Aliquots of NK-92 cells infected with vac-2DL4 were tested for their ability to lyse 721.221 (squares) or 221-G cells (triangles) in the presence of 5 μg/ml of the anti-class I mAb DX17 (open symbols) or control IgG1 (filled symbols). Similar results were obtained in two independent experiments.
Figure 3
Functional transfer of KIR2DL4 in NK-92 cells results in HLA-G–mediated inhibition. (A) Flow cytometry analysis of cell surface KIR2DL4 in NK-92 cells either uninfected, or infected with vac-2DL4 (15 PFU/cell). The cells were stained with either anti-2DL4 rabbit antiserum followed by FITC goat anti–rabbit IgG or with the mAb VV1-1G10 specific for a vaccinia cell surface protein. The histograms show log fluorescence of ungated cells. (B) Aliquots of uninfected or vac-2DL4–infected cells in A were tested for their ability to lyse 721.221 cells and 721.221 cells transfected with HLA-Cw*0401 and HLA-G, in a 4-h 51Cr-release assay. Lysis is shown for an E/T ratio of 5. Similar data was obtained at other E/T ratios and in five independent experiments. (C) Aliquots of NK-92 cells infected with vac-2DL4 were tested for their ability to lyse 721.221 (squares) or 221-G cells (triangles) in the presence of 5 μg/ml of the anti-class I mAb DX17 (open symbols) or control IgG1 (filled symbols). Similar results were obtained in two independent experiments.
Figure 4
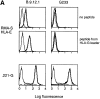
KIR2DL4-mediated recognition of HLA-G is reversed using a HLA-G–specific mAb that does not recognize HLA-E. (A) mAb G233 does not recognize HLA-E. RMAS-E cells were loaded with 500 μM of the HLA-G signal sequence–derived peptide VMAPRTLFL at 26°C and stained with either the anti-HLA reactive mAb B9.12.1 or the HLA-G specific mAb G233. The staining of 221-G cells using both antibodies is shown in the bottom panel. (B) Flow cytometry analysis of cell surface KIR2DL1 and KIR2DL4 in NK-92 cells infected with either vac-2DL1 or vac-2DL4. Cells were stained with mAb EB6 (specific for KIR2DL1), mAb VV1-1G10 (anti-vac), or anti-2DL4 rabbit antiserum followed by FITC-labeled species-specific secondary reagents. (C) Aliquots of infected cells expressing KIR2DL1 or KIR2DL4 were tested for their ability to lyse 221-Cw3, 221-Cw4, or 221-G target cells. The interaction between vac-2DL4 expressing NK-92 cells and those expressing 221-G was tested in the presence of either 5 μg/ml of isotype-matched control IgG2a (cIg) or HLA-G–specific mAb G233. Similar results were obtained in three independent experiments.
Figure 4
KIR2DL4-mediated recognition of HLA-G is reversed using a HLA-G–specific mAb that does not recognize HLA-E. (A) mAb G233 does not recognize HLA-E. RMAS-E cells were loaded with 500 μM of the HLA-G signal sequence–derived peptide VMAPRTLFL at 26°C and stained with either the anti-HLA reactive mAb B9.12.1 or the HLA-G specific mAb G233. The staining of 221-G cells using both antibodies is shown in the bottom panel. (B) Flow cytometry analysis of cell surface KIR2DL1 and KIR2DL4 in NK-92 cells infected with either vac-2DL1 or vac-2DL4. Cells were stained with mAb EB6 (specific for KIR2DL1), mAb VV1-1G10 (anti-vac), or anti-2DL4 rabbit antiserum followed by FITC-labeled species-specific secondary reagents. (C) Aliquots of infected cells expressing KIR2DL1 or KIR2DL4 were tested for their ability to lyse 221-Cw3, 221-Cw4, or 221-G target cells. The interaction between vac-2DL4 expressing NK-92 cells and those expressing 221-G was tested in the presence of either 5 μg/ml of isotype-matched control IgG2a (cIg) or HLA-G–specific mAb G233. Similar results were obtained in three independent experiments.
Figure 4
KIR2DL4-mediated recognition of HLA-G is reversed using a HLA-G–specific mAb that does not recognize HLA-E. (A) mAb G233 does not recognize HLA-E. RMAS-E cells were loaded with 500 μM of the HLA-G signal sequence–derived peptide VMAPRTLFL at 26°C and stained with either the anti-HLA reactive mAb B9.12.1 or the HLA-G specific mAb G233. The staining of 221-G cells using both antibodies is shown in the bottom panel. (B) Flow cytometry analysis of cell surface KIR2DL1 and KIR2DL4 in NK-92 cells infected with either vac-2DL1 or vac-2DL4. Cells were stained with mAb EB6 (specific for KIR2DL1), mAb VV1-1G10 (anti-vac), or anti-2DL4 rabbit antiserum followed by FITC-labeled species-specific secondary reagents. (C) Aliquots of infected cells expressing KIR2DL1 or KIR2DL4 were tested for their ability to lyse 221-Cw3, 221-Cw4, or 221-G target cells. The interaction between vac-2DL4 expressing NK-92 cells and those expressing 221-G was tested in the presence of either 5 μg/ml of isotype-matched control IgG2a (cIg) or HLA-G–specific mAb G233. Similar results were obtained in three independent experiments.
Similar articles
- Residues Met76 and Gln79 in HLA-G alpha1 domain involve in KIR2DL4 recognition.
Yan WH, Fan LA. Yan WH, et al. Cell Res. 2005 Mar;15(3):176-82. doi: 10.1038/sj.cr.7290283. Cell Res. 2005. PMID: 15780179 - Human KIR2DL5 is an inhibitory receptor expressed on the surface of NK and T lymphocyte subsets.
Estefanía E, Flores R, Gómez-Lozano N, Aguilar H, López-Botet M, Vilches C. Estefanía E, et al. J Immunol. 2007 Apr 1;178(7):4402-10. doi: 10.4049/jimmunol.178.7.4402. J Immunol. 2007. PMID: 17371997 - Rapid production of human KIR2DL4 extracellular domain and verification of its interaction with HLA-G.
Yu YR, Tian XH, Wang Y, Feng MF. Yu YR, et al. Biochemistry (Mosc). 2006;71 Suppl 1:S60-4, 4-5. doi: 10.1134/s0006297906130104. Biochemistry (Mosc). 2006. PMID: 16487070 - Natural killer cell-mediated recognition of human trophoblast.
Biassoni R, Bottino C, Millo R, Moretta L, Moretta A. Biassoni R, et al. Semin Cancer Biol. 1999 Feb;9(1):13-8. doi: 10.1006/scbi.1998.0108. Semin Cancer Biol. 1999. PMID: 10092546 Review. - Paired inhibitory and triggering NK cell receptors for HLA class I molecules.
López-Botet M, Bellón T, Llano M, Navarro F, García P, de Miguel M. López-Botet M, et al. Hum Immunol. 2000 Jan;61(1):7-17. doi: 10.1016/s0198-8859(99)00161-5. Hum Immunol. 2000. PMID: 10658973 Review.
Cited by
- Manifestations of immune tolerance in the human female reproductive tract.
Clark GF, Schust DJ. Clark GF, et al. Front Immunol. 2013 Feb 13;4:26. doi: 10.3389/fimmu.2013.00026. eCollection 2013. Front Immunol. 2013. PMID: 23407606 Free PMC article. - Stem cells and COVID-19: are the human amniotic cells a new hope for therapies against the SARS-CoV-2 virus?
Riedel RN, Pérez-Pérez A, Sánchez-Margalet V, Varone CL, Maymó JL. Riedel RN, et al. Stem Cell Res Ther. 2021 Mar 1;12(1):155. doi: 10.1186/s13287-021-02216-w. Stem Cell Res Ther. 2021. PMID: 33648582 Free PMC article. Review. - Matrix metalloproteinase-2 (MMP-2) generates soluble HLA-G1 by cell surface proteolytic shedding.
Rizzo R, Trentini A, Bortolotti D, Manfrinato MC, Rotola A, Castellazzi M, Melchiorri L, Di Luca D, Dallocchio F, Fainardi E, Bellini T. Rizzo R, et al. Mol Cell Biochem. 2013 Sep;381(1-2):243-55. doi: 10.1007/s11010-013-1708-5. Epub 2013 Jun 5. Mol Cell Biochem. 2013. PMID: 23737137 - KIR2DL4 (CD158d): An activation receptor for HLA-G.
Rajagopalan S, Long EO. Rajagopalan S, et al. Front Immunol. 2012 Aug 20;3:258. doi: 10.3389/fimmu.2012.00258. eCollection 2012. Front Immunol. 2012. PMID: 22934097 Free PMC article. - HLA-G and NK receptor are expressed in psoriatic skin: a possible pathway for regulating infiltrating T cells?
Aractingi S, Briand N, Le Danff C, Viguier M, Bachelez H, Michel L, Dubertret L, Carosella ED. Aractingi S, et al. Am J Pathol. 2001 Jul;159(1):71-7. doi: 10.1016/S0002-9440(10)61675-6. Am J Pathol. 2001. PMID: 11438456 Free PMC article.
References
- Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–393. - PubMed
- Braud VM, Allan DSJ, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. - PubMed
- Pazmany L, Mandelboim O, Vales-Gomez M, Davis DM, Reyburn HT, Strominger JL. Protection from natural killer cell-mediated lysis by HLA-G expression on target cells. Science. 1996;274:792–795. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials