Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States - PubMed (original) (raw)
Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States
D L Suarez et al. J Virol. 1999 May.
Abstract
The presence of low-pathogenic H7 avian influenza virus (AIV), which is associated with live-bird markets (LBM) in the Northeast United States, was first detected in 1994 and, despite efforts to eradicate the virus, surveillance of these markets has resulted in numerous isolations of H7 AIVs from several states from 1994 through 1998. The hemagglutinin, nonstructural, and matrix genes from representative H7 isolates from the LBM and elsewhere were sequenced, and the sequences were compared phylogenetically. The hemagglutinin gene of most LBM isolates examined appeared to have been the result of a single introduction of the hemagglutinin gene. Evidence for evolutionary changes were observed with three definable steps. The first isolate from 1994 had the amino acid threonine at the -2 position of the hemagglutinin cleavage site, which is the most commonly observed amino acid at this site for North American H7 AIVs. In January 1995 a new genotype with a proline at the -2 position was detected, and this genotype eventually became the predominant virus isolate. A third viral genotype, detected in November 1996, had an eight-amino-acid deletion within the putative receptor binding site. This viral genotype appeared to be the predominant isolate, although isolates with proline at the -2 position without the deletion were still observed in viruses from the last sampling date. Evidence for reassortment of multiple viral genes was evident. The combination of possible adaptive evolution of the virus and reassortment with different influenza virus genes makes it difficult to determine the risk of pathogenesis of this group of H7 AIVs.
Figures
FIG. 1
Amino acid sequence comparison of the HA1 coding region of three North American H7 isolates selected to display unique structural features of the HA1 coding region. The amino acids which have been shown by molecular modeling to correspond to the receptor binding site identified in the H3 structure are in boldface and are underlined. The serine insertion in the quail isolate between positions 136 and 137 is indicated by the dash at that position in the two 1996 isolates. The amino acid numbering system is based upon the consensus H7 amino acid sequence. The cleavage site separating the HA1 and HA2 is indicated by the gap.
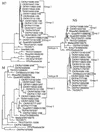
FIG. 2
Phylogenetic trees from the H7 hemagglutinin (H7) subtype, NS subtypes (groups A and B), and the M gene from H7 isolates from the LBMs in the Northeast United States and other representative AIV isolates are presented. All trees were generated with PAUP 3.1 computer program, are the result of 100 bootstrap replicates, and are midpoint rooted. Branch lengths are included on each tree. The H7 LBM isolates are grouped (1 to 4 for the H7 and NS trees and 1 to 3 for the M tree) according to their associations on the tree. Abbreviations: CK, chicken; TK, turkey; GF, guinea fowl; and FPV, Fowl plague virus. Standard two-letter abbreviations are used for states in the United States.
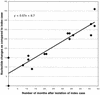
FIG. 3
Comparison of the evolutionary rate of the HA1 segment of the hemagglutinin gene from H7 AIV isolates from the LBMs of the Northeastern United States. The number of nucleotide changes from the earliest H7 isolate, A/CK/NJ/15086-3/94, are given on the y axis, and the number of months after each virus was isolated compared to the index case is given on the x axis. Linear regression analysis was performed to determine the evolutionary rate, and the equation for the line is indicated.
Similar articles
- Molecular and biological characteristics of H5 and H7 avian influenza viruses in live-bird markets of the northeastern United States, 1994-2001.
Senne DA, Suarez DL, Pedersen JC, Panigrahy B. Senne DA, et al. Avian Dis. 2003;47(3 Suppl):898-904. doi: 10.1637/0005-2086-47.s3.898. Avian Dis. 2003. PMID: 14575083 - Avian influenza virus subtypes inside and outside the live bird markets, 1993-2000: a spatial and temporal relationship.
Panigrahy B, Senne DA, Pedersen JC. Panigrahy B, et al. Avian Dis. 2002 Apr-Jun;46(2):298-307. doi: 10.1637/0005-2086(2002)046[0298:AIVSIA]2.0.CO;2. Avian Dis. 2002. PMID: 12061638 - Molecular analysis of H7 avian influenza viruses from Australia and New Zealand: genetic diversity and relationships from 1976 to 2007.
Bulach D, Halpin R, Spiro D, Pomeroy L, Janies D, Boyle DB. Bulach D, et al. J Virol. 2010 Oct;84(19):9957-66. doi: 10.1128/JVI.00930-10. Epub 2010 Jul 28. J Virol. 2010. PMID: 20668069 Free PMC article. - Avian influenza viruses in New Zealand wild birds, with an emphasis on subtypes H5 and H7: Their distinctive epidemiology and genomic properties.
Stanislawek WL, Tana T, Rawdon TG, Cork SC, Chen K, Fatoyinbo H, Cogger N, Webby RJ, Webster RG, Joyce M, Tuboltsev MA, Orr D, Ohneiser S, Watts J, Riegen AC, McDougall M, Klee D, O'Keefe JS. Stanislawek WL, et al. PLoS One. 2024 Jun 3;19(6):e0303756. doi: 10.1371/journal.pone.0303756. eCollection 2024. PLoS One. 2024. PMID: 38829903 Free PMC article. - Advances in the molecular based techniques for the diagnosis and characterization of avian influenza virus infections.
Pasick J. Pasick J. Transbound Emerg Dis. 2008 Oct;55(8):329-38. doi: 10.1111/j.1865-1682.2008.01047.x. Transbound Emerg Dis. 2008. PMID: 18786072 Review.
Cited by
- Susceptibility of Chickens to Low Pathogenic Avian Influenza (LPAI) Viruses of Wild Bird- and Poultry-Associated Subtypes.
Bergervoet SA, Germeraad EA, Alders M, Roose MM, Engelsma MY, Heutink R, Bouwstra R, Fouchier RAM, Beerens N. Bergervoet SA, et al. Viruses. 2019 Oct 31;11(11):1010. doi: 10.3390/v11111010. Viruses. 2019. PMID: 31683727 Free PMC article. - Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site.
Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Yang H, et al. PLoS Pathog. 2010 Sep 2;6(9):e1001081. doi: 10.1371/journal.ppat.1001081. PLoS Pathog. 2010. PMID: 20824086 Free PMC article. - Continued circulation in China of highly pathogenic avian influenza viruses encoding the hemagglutinin gene associated with the 1997 H5N1 outbreak in poultry and humans.
Cauthen AN, Swayne DE, Schultz-Cherry S, Perdue ML, Suarez DL. Cauthen AN, et al. J Virol. 2000 Jul;74(14):6592-9. doi: 10.1128/jvi.74.14.6592-6599.2000. J Virol. 2000. PMID: 10864673 Free PMC article. - Diagnostic approach for differentiating infected from vaccinated poultry on the basis of antibodies to NS1, the nonstructural protein of influenza A virus.
Tumpey TM, Alvarez R, Swayne DE, Suarez DL. Tumpey TM, et al. J Clin Microbiol. 2005 Feb;43(2):676-83. doi: 10.1128/JCM.43.2.676-683.2005. J Clin Microbiol. 2005. PMID: 15695663 Free PMC article. - Pathobiology and transmission of highly and low pathogenic avian influenza viruses in European quail (Coturnix c. coturnix).
Bertran K, Dolz R, Busquets N, Gamino V, Vergara-Alert J, Chaves AJ, Ramis A, Abad FX, Höfle U, Majó N. Bertran K, et al. Vet Res. 2013 Mar 28;44(1):23. doi: 10.1186/1297-9716-44-23. Vet Res. 2013. PMID: 23537387 Free PMC article.
References
- Banks J, Speidel E, Alexander D J. Characterization of an avian influenza A virus isolated from a human—is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch Virol. 1998;143:781–787. - PubMed
- Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. - PubMed
- Bosch F X, Garten W, Klenk H D, Rott R. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology. 1981;113:725–735. - PubMed
- Daly J M, Lai A C, Binns M M, Chambers T M, Barrandeguy M, Mumford J A. Antigenic and genetic evolution of equine H3N8 influenza A viruses. J Gen Virol. 1996;77:661–671. - PubMed
- Davidson W R, Yoder H W, Brugh M, Nettles V F. Serological monitoring of Eastern Wild Turkeys for antibodies to Mycoplasma spp. and avian influenza viruses. J Wildl Dis. 1988;24:348–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical