Genetic studies exposing the splicing events involved in herpes simplex virus type 1 latency-associated transcript production during lytic and latent infection - PubMed (original) (raw)
Genetic studies exposing the splicing events involved in herpes simplex virus type 1 latency-associated transcript production during lytic and latent infection
M R Alvira et al. J Virol. 1999 May.
Abstract
Herpes simplex virus type 1 (HSV-1) establishes latency in sensory neurons, a state in which the viral lytic genes are silenced and only the latency locus is transcriptionally active, producing the 2. 0- and 1.5-kb latency-associated transcripts (LATs). Previous experimental evidence indicates that the LATs are stable introns, and it has been reported that LAT formation is abolished by debilitating substitution mutations in the predicted splice sites during lytic infection but not latency (J. L. Arthur et al., J. Gen. Virol. 79:107-116, 1998). We have independently studied a set of deletion mutations to explore the roles of the proposed splice sites during lytic and latent infection. HSV-1 mutant viruses missing the invariant intron-terminal 5'-G(T/C) or 3'-AG dinucleotides were analyzed for LAT formation during lytic infection in vitro, when only the 2-kb LAT is produced, and during latency in mouse trigeminal ganglia, where both LATs are expressed. Northern blot analysis of total RNAs from different productively infected cell lines showed that the lytic (2-kb) LAT was not expressed by the various splice site deletion mutants. In vivo studies using a mouse eye model of latency similarly showed that the latent (2- and 1. 5-kb) LATs were not expressed by the mutants. PCR analysis with primers flanking the LAT sequence revealed the expected splice junction for LAT excision in RNA from sensory neurons latently infected with wild-type but not mutant virus. Using a virus mutant deleted in the splicing signals flanking the 556-bp region of LAT whose absence distinguishes the 1.5- and 2-kb LATs, we observed selective elimination of 1.5-kb LAT expression in latency, supporting previous suggestions that the internal region is removed by splicing. Taken together, these results demonstrate that the 2-kb LAT is formed during both lytic and latent infection by splicing at the predicted splice sites and that an additional splicing event is involved in the latency-restricted production of the 1.5-kb LAT. We have also mapped the 3' end of the lytic 2-kb LAT and discuss our results in the context of previous models addressing the unusual stability of the LATs.
Figures
FIG. 1
Schematic representation of the LATs of HSV-1 depicting the locations of probes and splice site mutations. (A) The LAT genes in the inverted repeats flanking UL and the positions of relevant restriction sites based on the published sequence of HSV-1 strain 17+ (44). (B) The minor 8.3-kb (mLAT) and major 2.0- and 1.5-kb LATs, transcribed from left to right, overlap with the ICP0 transcript transcribed off of the opposite strand. The positions of the two LAT promoters, LAP1 and LAP2, are indicated. Also diagrammed are the start site (nt 118801) and direction of transcription. Crosses over the ICP0 mRNA represent the region of complementarity to the major LATs that is read out of frame in the splice acceptor mutant; the arrowhead at the end of the mRNA indicates the direction of transcription. The positions of the canonical splice donor (SD) and splice acceptor (SA) sites potentially involved in the generation of the major LATs (2.0 and 1.5 kb) are also shown. (C) Locations of the probes used for Southern and Northern blot analysis. S, _Sph_I; B, _Bbs_I; A, _Alw_NI; Sa, _Sal_I; M, _Mlu_I. (D) Alignment of the LAT splicing signals with eukaryotic consensus sites. The splice donor and splice acceptor consensus sequences are shown in boxes representing two exons (shaded) and an intron (open); the frequencies of these consensus nucleotides among vertebrate splice site sequences are listed underneath as percentages (35). The canonical splicing signals bounding the 2-kb LAT and the presumed internal intron (1.5-kb LAT) are shown below in alignment with the consensus sequences; the invariant dinucleotides at the intron-terminal positions are in boldface. The bases deleted in the splice site mutant viruses of this study are shown in lowercase. Py, pyrimidine; N, any nucleotide.
FIG. 2
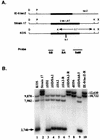
Southern blot analysis of virus recombinants deleted for LAT splice donor/acceptor sites. (A) LAT locus of the ICP0 mutant of strain 17 in which the ICP0 coding sequence was replaced by the lacZ gene, removing the 3′ terminus of the LAT intron (9). Unique _Kpn_I (K) and _Pml_I (P) restriction enzyme sites within the _Dra_I (D) and _Xho_I (X) fragments of each parental virus that were used to distinguish KOS from strain 17 sequences in the recombinant virus mutants are noted. The KOS LAT intron sequence also contains a 96-bp repeat (indicated as 0.1 at the Δ) resulting from a different copy number of a 16-nt repeat element; strain 17 syn+ contains 9 copies, while KOS contains 15 copies (64). Underneath are the locations of the radiolabeled strain 17 probes (SB, BA, and SaM) used simultaneously for Southern blot analysis of the _Kpn_I digests (see also Fig. 1C and Materials and Methods). (B) Southern blot analysis of the SD/SA mutant viruses. Note that KOS parental virus (lane 1) contains three bands whereas strain 17 and the rescuant IE-0:_lacZ_-R (lanes 2 and 10, respectively) contain only two slower-migrating specific bands that are larger by ca. 2.7-kb sequence found only in KOS. Lanes 3 to 5 and 9 show the presence of KOS sequences in the 2-kb LAT SD/SA mutants and in the 1.5-kb LAT SD/SA mutant, respectively; lanes 6 and 7 show the virus rescuant patterns; lane 8 shows a faster-migrating strain 17 doublet due to the removal of the 0.5-kb internal intron sequences. Sizes are indicated in nucleotides.
FIG. 3
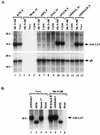
Northern blot analysis of total RNAs isolated from productively infected cells in culture. (A) LAT expression from viruses with mutations in the external splice site signals at 8 or 14 h p.i. (indicated by virus names at the top) as visualized by hybridization with LAT-specific probe SS (Fig. 1C). The lower panel shows the stripped blot rehybridized with an HSV-1 gB-specific probe as a control. Virus names ending with −R designate rescued mutants. (B) LAT expression at 12 h p.i., using the SS probe, by a virus with mutated internal splice site signals (lanes 1 and 6) compared with expression from control viruses, including _dl_556, which has an exact deletion of the internal intron and is seen to produce 1.5-kb LAT under lytic conditions (lanes 2 and 7) instead of 2-kb LAT.
FIG. 4
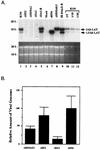
In vivo analysis of LAT expression by the mutant splice site recombinants. (A) Northern blot analysis of LAT expression in latently infected trigeminal ganglia. The probe used for LAT detection was the same as in Fig. 2, i.e., a mixture of probes SB, BA, and SaM (see also Fig. 1). Lanes 10 to 12 show 5-, 10-, and 20-fold dilutions, respectively, of the wt KOS RNA loaded in lane 1 to assess the sensitivity of the experiment. The _dl_556 mutant (lane 7) produces a LAT species that is 556 nt smaller than wild-type 2-kb LAT. The lower panel displays the ethidium bromide-stained gel prior to transfer demonstrating equal loading of the samples. (B) PCR analysis of latent viral genomes. Quantitative PCR analysis was performed on DNA samples from latently infected trigeminal ganglia, and the results were converted to numbers representing relative levels of latency based on HSV genome abundance (see Materials and Methods for a description of the procedures). DNA from _dl_556-infected ganglia yielded the highest mean value of triplicate determinations, and this value was arbitrarily set as the standard (100%).
FIG. 5
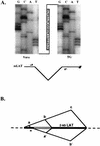
Exonic product of splicing between the canonical splice sites bounding 2-kb LAT in lytic and latent infections detected by RT-PCR with a summary of splicing events in latent infections. (A) RT-PCR analysis of the exon junction resulting from splicing of the 2-kb LAT within spliced mLAT. RNAs were isolated from strain 17-infected Vero cells at 12 h p.i. and from latently infected trigeminal ganglia (TG). Aliquots were reverse transcribed from an mLAT-specific primer downstream of the 2-kb LAT region, and the products were amplified by PCR with mLAT-specific primers narrowly flanking the 2-kb LAT sequence, as diagrammed. Sequencing of the single RT-PCR product from each reaction demonstrated the joining in both situations of mLAT sequences at the canonical splicing signals bounding the major LATs. The autoradiographs show the sequences through the splice junction. (B) Summary of splicing events in latency. The 1.5-kb LAT sequence is diagrammed as two black boxes separated by an open box representing the internal intron, which together represent the 2-kb LAT sequence; flanking mLAT sequences are shown as dotted lines. Splice joints identified by RT-PCR are illustrated above and below the diagram as intersecting diagonal lines connecting the two sides of the joint. *, cryptic splice acceptor previously seen in minigene-transfected but not virus-infected cells.
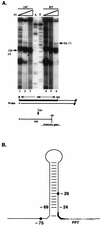
FIG. 6
S1 nuclease protection mapping of the 3′ ends of lytic 2-kb LAT and positions of the ends relative to previously proposed features. (A) S1 analysis of the 3′ ends of the 2-kb LAT. Increasing amounts of S1 enzyme (10 to 300 U) were used for digestions at 14°C (14C) and room temperature (RT). RNA was isolated from strain 17-infected SK-N-SH cells at 12 h p.i. The sketch of 3′-end-labeled probe below the autoradiograph indicates the position of the label (*) and the distance to the external splice acceptor site. The main protected bands on the autoradiograph at the highest enzyme concentrations (lanes 3 and 6) migrate at 123 to 124 nt, as determined by alignment with the A/T sequencing ladder. A product potentially extending to the splice acceptor (SA) is indicated. The main digestion products are sketched at the bottom, with the final protected nucleotides shown in capitals. (B) Proposed stem-loop structure in the 3′ region of LAT. All base pairs proposed by Krummenacher et al. (37) are represented, including a potentially unstable A:T pair at the base of the stem below two unpaired positions. The polypyrimidine tract (PPT) is shaded, and the alternately proposed branch points at −29 and −75 are marked by dots. The drawing illustrates that position −24, representing a major 3′ end of LAT, is located at the bottom of the stable portion of the hairpin and that the potential branch point at position −29 is inside the hairpin.
Similar articles
- Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections.
Chen X, Schmidt MC, Goins WF, Glorioso JC. Chen X, et al. J Virol. 1995 Dec;69(12):7899-908. doi: 10.1128/JVI.69.12.7899-7908.1995. J Virol. 1995. PMID: 7494302 Free PMC article. - A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation.
Kennedy PG, Rovnak J, Badani H, Cohrs RJ. Kennedy PG, et al. J Gen Virol. 2015 Jul;96(Pt 7):1581-602. doi: 10.1099/vir.0.000128. Epub 2015 Mar 20. J Gen Virol. 2015. PMID: 25794504 Free PMC article. Review. - The latency-associated gene of herpes simplex virus type 1 (HSV-1) interferes with superinfection by HSV-1.
Mador N, Panet A, Steiner I. Mador N, et al. J Neurovirol. 2002 Dec;8 Suppl 2:97-102. doi: 10.1080/13550280290167920. J Neurovirol. 2002. PMID: 12491159 Review.
Cited by
- Analysis of protein expression from within the region encoding the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1.
Lock M, Miller C, Fraser NW. Lock M, et al. J Virol. 2001 Apr;75(7):3413-26. doi: 10.1128/JVI.75.7.3413-3426.2001. J Virol. 2001. PMID: 11238867 Free PMC article. - Herpes simplex virus type 1 latently infected neurons differentially express latency-associated and ICP0 transcripts.
Maillet S, Naas T, Crepin S, Roque-Afonso AM, Lafay F, Efstathiou S, Labetoulle M. Maillet S, et al. J Virol. 2006 Sep;80(18):9310-21. doi: 10.1128/JVI.02615-05. J Virol. 2006. PMID: 16940542 Free PMC article. - Construction of a herpes simplex virus type 1 mutant with only a three-nucleotide change in the branchpoint region of the latency-associated transcript (LAT) and the stability of its two-kilobase LAT intron.
Ng AK, Block TM, Aiamkitsumrit B, Wang M, Clementi E, Wu TT, Taylor JM, Su YH. Ng AK, et al. J Virol. 2004 Nov;78(22):12097-106. doi: 10.1128/JVI.78.22.12097-12106.2004. J Virol. 2004. PMID: 15507596 Free PMC article. - Herpes simplex virus type 1 promoter activity during latency establishment, maintenance, and reactivation in primary dorsal root neurons in vitro.
Arthur JL, Scarpini CG, Connor V, Lachmann RH, Tolkovsky AM, Efstathiou S. Arthur JL, et al. J Virol. 2001 Apr;75(8):3885-95. doi: 10.1128/JVI.75.8.3885-3895.2001. J Virol. 2001. PMID: 11264377 Free PMC article. - Role of activating transcription factor 3 in the synthesis of latency-associated transcript and maintenance of herpes simplex virus 1 in latent state in ganglia.
Shu M, Du T, Zhou G, Roizman B. Shu M, et al. Proc Natl Acad Sci U S A. 2015 Sep 29;112(39):E5420-6. doi: 10.1073/pnas.1515369112. Epub 2015 Aug 24. Proc Natl Acad Sci U S A. 2015. PMID: 26305977 Free PMC article.
References
- Aebi M, Hornig H, Padgett R A, Reiser J, Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986;47:555–565. - PubMed
- Arthur J L, Everett R, Brierley I, Efstathiou S. Disruption of the 5′ and 3′ splice sites flanking the major latency-associated transcripts of herpes simplex virus type 1: evidence for alternate splicing in lytic and latent infections. J Gen Virol. 1998;79:107–116. - PubMed
- Berk A J. Characterization of RNA molecules by S1 nuclease analysis. Methods Enzymol. 1989;180:335–347. - PubMed
- Block T M, Hill J M. The latency associated transcripts (LAT) of herpes simplex virus: still no end in sight. J Neurovirol. 1997;3:313–321. - PubMed
- Brown S M, Ritchie D A, Subak-Sharpe J H. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973;18:329–346. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials