Design of 5' untranslated sequences in retroviral vectors developed for medical use - PubMed (original) (raw)
Design of 5' untranslated sequences in retroviral vectors developed for medical use
M Hildinger et al. J Virol. 1999 May.
Abstract
Utilizing genetic modules of simple retroviruses, we have developed a novel generation of gene transfer vectors with improved therapeutic potential. In the 5' untranslated "leader" sequences, all AUG codons which may aberrantly initiate translation and all viral coding sequences were removed. Thus, the probability of expressing unwanted peptides and the potential for homologous recombination with retroviral genes were largely reduced, and the cloning capacity was increased. The transgene was inserted to replace the viral gag sequences, and a new minimal splice acceptor was introduced, resulting in increased expression with all genes tested (those coding for human multidrug resistance 1 and enhanced green fluorescent protein, as well as the lacZ gene). These vectors may represent attractive tools for human gene therapy, because they increase the efficiency of transgene expression and may also increase safety in medical applications.
Figures
FIG. 1
Design of leader sequences. A schematic representation of the 5′ untranslated sequences used in retroviral vectors containing viral gene fragments (first set) and the novel design analyzed in this study (second set) is shown. Ψ, packaging signal; SD, splice donor; SA, splice acceptor. Open boxes indicate the viral gag gene fragment (G+), and shaded boxes indicate the viral pol gene fragment (P+). Potential AUG start codons preceding the cDNA are indicated by open circles, and the AUG of the inserted cDNA is indicated by a solid circle. G+SD−, G+ vector with destroyed splice donor; GRP+, gag replacement with P+ present; GR, gag replacement; GRSD−, GR vector with destroyed splice donor; GRS, gag replacement with SA oligonucleotide. GRSΔ270, GRS leader with destroyed AUG at position +270.
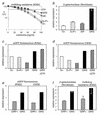
FIG. 2
The novel GRS leader (gag replacement with splice) increases expression from retroviral vectors. (a) The expression of multidrug resistance in K562 cells transduced with MDR1 vectors is improved with GR leader sequences. Uncloned mass cultures of cells transduced with the vectors as described in Materials and Methods were plated in colony assays containing increasing doses of the cytotoxic agent colchicine, which is recognized by the MDR1-encoded drug efflux pump. The surviving colonies were microscopically counted. Data are expressed as relative cloning efficiency of colony-forming cells (CFC) with respect to the unselected control. The novel leaders GR and, even better, GRS significantly increase the ID50 of the drug, which is 3 ng/ml for untransduced cells. Shown are mean values of two separate experiments, with each culture plated for nine determinations. Most standard deviations were below 15%; only at doses of >60 ng of colchicine/ml were standard deviations up to 26%. Differences between vectors GRS and G+ are highly significant (P < 0.001). Regression analysis revealed the linearity of the curves (_r_ values of >0.995). The following monocistronic vectors described in Materials and Methods were used: SF1m (G+), SF6m (G+P+), SF11m (GR), and SF71m (GRS). (b) The GRS leader improves expression of β-galactosidase from vectors containing the _lacZ_-Neor fusion gene. Transduced NIH-3T3-based fibroblasts were selected as a mass culture with G418 at 400 μg/ml, and β-galactosidase activity was determined in cell extracts, as described in Materials and Methods. Data are expressed as relative protein expression with respect to cells transduced with SF1K containing the G+ leader. Standard deviations of three experiments were below 20%. The following monocistronic vectors described in Materials and Methods were used: SF1K (G+), SF6K (G+P+), SF11K (GR), and SF71K (GRS). (c and d) Increased expression of eGFP with the GRS leader in human hematopoietic (c) and lymphoid (d) cells. The destruction of the final residual AUG at +270 slightly increases expression from the GRS leader (GRSΔ270). Data represent the mean fluorescence of unselected transduced cells measured by flow cytometry as described in Materials and Methods, expressed as fold increase with respect to cells transduced with SF1γ containing the G+ leader. A total of 200,000 cells were measured per experiment. The data were reproduced in two further sets of experiments, with standard deviations of <11%. Differences between GRS vectors and vectors G+, G+P+, and GR were significant (P < 0.005). The following monocistronic vectors described in Materials and Methods were used: SF1γ (G+), SF6γ (G+P+), SF11γ (GR), SF71γ (GRS), and SF91γ (GRSΔ270). (e and f) The splice acceptor oligonucleotide contained in the GRS leader is superior to the pol gene fragment (P+) for protein expression with all genes tested. (e) Increase in mean fluorescence with respect to the G+ vector. Data were accumulated and expressed as described for panels c and d. The following monocistronic vectors described in Materials and Methods were used: SF1γ (G+), SF61γ (GRP+), and SF71γ (GRS). (f) The GRS vector is superior to the GRP+ vector in expression of MDR1. Data are expressed with respect to those achieved with the G+ vector, shown for cloning efficiency in the presence of 100 ng of colchicine/ml. Mean values of nine determinations with error bars representing standard deviations are presented, showing significant differences between vectors GRS and GRP+ (P < 0.005). Data were reproduced in a second experiment. The following monocistronic vectors described in Materials and Methods were used: SF1m (G+), SF61m (GRP+), and SF71m (GRS). Assay procedures were as described for panel a.
FIG. 3
Efficient splicing of 5′ untranslated leader sequences expressed from GRS vectors. RT-PCR was performed with RNA extracted from mass cultures of K562 cells transduced with SF1m (G+ leader) and SF71m (GRS leader) using primers annealing upstream of the retroviral splice donor and downstream of the start codon of the MDR1 cDNA. Sizes of the unspliced (U) RNA are 1,214 and 833 bp for SF1m (G+) and SF71m (GRS), respectively. The sizes of the spliced (S) RNA are 376 bp and 364 bp for SF1m (G+) and SF71m (GRS), respectively. The identity of the spliced RNA was confirmed by sequencing of subcloned RT-PCR products (Fig. 4). We also observed previously unknown, minor alternative splice (aS) products of both leader sequences whose origins remain to be determined. M, molecular weight marker; no, untransduced cells.
FIG. 4
Sequence of the GRSΔ270 leader (gag replacement with splice) based on MESV, starting from the cap site and ending with the ATG of the cDNA (circled). The primer binding site (PBS) and recognition sites for endonucleases _Kpn_I, _Msc_I, _Bgl_II, _Xma_III, and _Pst_I are indicated above the sequence. The splice donor (SD) and splice acceptor (SA) are boxed. The mutated AUG at position 270 and the SAO are shown in lowercase, and the stretch of the SAO with continuous sequence homology to pol is shown in boldface. Sequences contained in the major splice product shown in Fig. 3 are shown in italics. Putative non-AUG start codons are underlined, including the partially degenerated start codon for the glyco-Gag at position 366 (see Discussion). The sequence was obtained from plasmid pSF91 and by subcloning and repetitive sequencing of the major splice product of the RT-PCR shown in Fig. 3. Asterisks delineate sequence deviations (three A-G switches and one T insertion) compared to the published sequence of _dl587_rev (12). These are present in MESV and all derived leader variants analyzed in this study, including pSF1 (GenBank accession no. AJ224005).
Similar articles
- The position of the AUG start codon in MFG-based γ-retroviral vectors has a dramatic effect on translation-dependent protein expression.
Frankel TL, Zhang L, Burns WR, Zheng Z, Morgan RA. Frankel TL, et al. J Gene Med. 2011 Sep;13(9):478-86. doi: 10.1002/jgm.1599. J Gene Med. 2011. PMID: 21796743 - FMEV vectors: both retroviral long terminal repeat and leader are important for high expression in transduced hematopoietic cells.
Hildinger M, Eckert HG, Schilz AJ, John J, Ostertag W, Baum C. Hildinger M, et al. Gene Ther. 1998 Nov;5(11):1575-9. doi: 10.1038/sj.gt.3300759. Gene Ther. 1998. PMID: 9930313 - Retroviral vectors produced in the cytoplasmic vaccinia virus system transduce intron-containing genes.
Konetschny C, Holzer GW, Falkner FG. Konetschny C, et al. J Virol. 2002 Feb;76(3):1236-43. doi: 10.1128/jvi.76.3.1236-1243.2002. J Virol. 2002. PMID: 11773399 Free PMC article. - Mutagenesis by retroviral transgene insertion: risk assessment and potential alternatives.
Baum C, Fehse B. Baum C, et al. Curr Opin Mol Ther. 2003 Oct;5(5):458-62. Curr Opin Mol Ther. 2003. PMID: 14601513 Review. - Retroviral vectors for analysis of viral mutagenesis and recombination.
Rawson JM, Mansky LM. Rawson JM, et al. Viruses. 2014 Sep 24;6(9):3612-42. doi: 10.3390/v6093612. Viruses. 2014. PMID: 25254386 Free PMC article. Review.
Cited by
- Pbx1 is required for adult subventricular zone neurogenesis.
Grebbin BM, Hau AC, Groß A, Anders-Maurer M, Schramm J, Koss M, Wille C, Mittelbronn M, Selleri L, Schulte D. Grebbin BM, et al. Development. 2016 Jul 1;143(13):2281-91. doi: 10.1242/dev.128033. Epub 2016 May 25. Development. 2016. PMID: 27226325 Free PMC article. - Evaluation of different protocols for gene transfer into non-obese diabetes/severe combined immunodeficiency disease mouse repopulating cells.
Ebeling P, Bach P, Sorg U, Schneider A, Trarbach T, Dilloo D, Hanenberg H, Niesert S, Seeber S, Moritz T, Flasshove M. Ebeling P, et al. J Cancer Res Clin Oncol. 2007 Mar;133(3):199-209. doi: 10.1007/s00432-006-0158-9. Epub 2006 Oct 20. J Cancer Res Clin Oncol. 2007. PMID: 17053889 - Rescue of pyruvate kinase deficiency in mice by gene therapy using the human isoenzyme.
Meza NW, Alonso-Ferrero ME, Navarro S, Quintana-Bustamante O, Valeri A, Garcia-Gomez M, Bueren JA, Bautista JM, Segovia JC. Meza NW, et al. Mol Ther. 2009 Dec;17(12):2000-9. doi: 10.1038/mt.2009.200. Epub 2009 Sep 15. Mol Ther. 2009. PMID: 19755962 Free PMC article. - Nanobody-based CD38-specific heavy chain antibodies induce killing of multiple myeloma and other hematological malignancies.
Schriewer L, Schütze K, Petry K, Hambach J, Fumey W, Koenigsdorf J, Baum N, Menzel S, Rissiek B, Riecken K, Fehse B, Röckendorf JL, Schmid J, Albrecht B, Pinnschmidt H, Ayuk F, Kröger N, Binder M, Schuch G, Hansen T, Haag F, Adam G, Koch-Nolte F, Bannas P. Schriewer L, et al. Theranostics. 2020 Feb 3;10(6):2645-2658. doi: 10.7150/thno.38533. eCollection 2020. Theranostics. 2020. PMID: 32194826 Free PMC article. - Phosphorylation of murine SAMHD1 regulates its antiretroviral activity.
Wittmann S, Behrendt R, Eissmann K, Volkmann B, Thomas D, Ebert T, Cribier A, Benkirane M, Hornung V, Bouzas NF, Gramberg T. Wittmann S, et al. Retrovirology. 2015 Dec 15;12:103. doi: 10.1186/s12977-015-0229-6. Retrovirology. 2015. PMID: 26667483 Free PMC article.
References
- Anderson W F, McGarrity G J, Moen R C. Report to the NIH recombinant DNA advisory committee on murine replication competent retrovirus (RCR) assay. Hum Gene Ther. 1993;4:311–321. - PubMed
- Baum C, Stocking C, Wagener T, Eckert H-G, Ostertag W. Gene transfer and transgene expression in hematopoietic cells. In: Strauss M, Barranger J A, editors. Concepts in gene therapy. Berlin, Germany: De Gruyter; 1997. pp. 233–266.
- Baum C, Itoh K, Meyer J, Laker C, Ito Y, Ostertag W. The potent enhancer activity of the polycythemic strain of spleen focus-forming virus in hematopoietic cells is governed by a binding site for Sp1 in the upstream control region and by a unique enhancer core motif, creating an exclusive target for PEBP/CBF. J Virol. 1997;71:6323–6331. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources