Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs - PubMed (original) (raw)
Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs
K Tomonaga et al. J Virol. 1999 May.
Abstract
To develop a better understanding of the interaction between retroviruses and their hosts, we have investigated the polymorphism in endogenous murine leukemia proviruses (MLVs). We used genomic libraries of wild mouse DNAs and PCR to analyze genetic variation in the proviruses found in wild mouse species, including Mus musculus (M. m. castaneus, M. m. musculus, M. m. molossinus, and M. m. domesticus), Mus spretus, and Mus spicelegus, as well as some inbred laboratory strains. In this analysis, we detected several unique forms of sequence organization in the U3 regions of the long terminal repeats of these proviruses. The distribution of the proviruses with unique U3 structures demonstrated that xenotropic MLV-related proviruses were present only in M. musculus subspecies, while polytropic MLV-related proviruses were found in both M. musculus and M. spretus. Furthermore, one unique provirus from M. spicelegus was found to be equidistant from ecotropic provirus and nonecotropic provirus by phylogenetic analysis. This provirus, termed HEMV, was thus likely to be related to the common ancestor of these MLVs. Moreover, an ancestral type of polytropic MLV-related provirus was detected in M. spretus species. Despite their "ancestral" phylogenetic position, proviruses of these types are not widespread in mice, implying more-recent spread by infection rather than inheritance. These results imply that recent evolution of these proviruses involved alternating periods of replication as virus and residence in the germ line.
Figures
FIG. 1
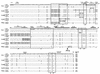
Structure of the MLV LTR. The unique sequences in the U3 region are indicated. The locations of primers and probes used in this study are also shown. The numbering of the boxed regions corresponds to that in our previous study (55). UCR, upstream conserved region (17); DR, direct repeat.
FIG. 2
U3 regions of xenotropic MLV-related proviruses. U3 sequences were cloned from M. musculus DNAs following amplification with the KS-50 and Uniltr-4 primers. Sequences which were potentially reactive the Xltr probe and which lacked the 190-bp insert are aligned with those of several previously sequenced provirus isolates: NFS (NFS-Th-1) (28), CWM (CWM-S-5X) (35), Bxv1 (23), Xmv44 (52), MX30 (49), Xmv28 (6), MX27, and MX33 (49). The sequence of NFS xenotropic provirus was used as a standard. Dots indicate nucleotide identity. Dashes indicate absence of a nucleotide. Direct repeats and unique sequences present in the proviruses are boxed. Potential enhancer sequence regions and transcriptional factor binding sites are indicated by the shaded and black bars, respectively. Locations of two promoter-associated motifs, the CAT and TATA boxes, are also indicated. The position of the 190-bp insertion in polytropic virus-related proviruses, MX27 and MX33, is shown by an arrow. The sequences of oligonucleotide probes used for unblotting analysis are underlined. The conserved _Pst_I recognition site is also shown. The size of the U3 region of each provirus is indicated at the end of the sequence in parentheses.
FIG. 3
Alignment of U3 sequences of polytropic MLV-related proviruses. Sequences of polytropic MLV-related U3 regions cloned from M. musculus and M. spretus DNAs as described in the previous legend are aligned with those of polytropic (MX27) and modified polytropic (MX33) proviruses (49). The MX27 sequence was used as a standard. Dots indicate nucleotide identity. Dashes indicate absence of a nucleotide. Direct repeats and unique sequences present in the proviruses are boxed (55). The 190-bp inserted region is also boxed. Potential enhancer sequence regions are indicated by the shaded bar. Locations of two promoter-associated motifs, the CAT and TATA boxes, are also indicated. The sequences of oligonucleotide probes are underlined. The conserved _Pst_I recognition site is also shown. The size of the U3 region of each provirus is indicated at the end of the sequence in parentheses.
FIG. 4
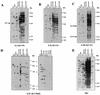
Distribution of xenotropic MLV-related proviruses in wild mice. Analysis of _Pvu_II-digested mouse DNAs was performed by hybridization of dried agarose gels with an X-I-specific oligonucleotide probe (KT-55) (A), an X-II-specific oligonucleotide probe (KT-51) (B), an X-III-specific oligonucleotide probe (KT-53) (C), and the X-IV-reactive oligonucleotide probe (KT-59/60) (D). Lanes (a to d contain laboratory [lab.] strains): a, AKR/J; b, HRS/J; c, C3H/HeJ; d, C57BL/6J; e, CAST/Ei (M. m. castaneus) (m.m.cas.); f, CZECH II/Ei (M. m. musculus) (m.m.mus.); g, MOLC/Rk (M. m. molossinus); h, MOLF/Ei (M. m. molossinus); i, MOLG/Dn (M. m. molossinus) (m.m.mol.); j, WSB/Ei (M. m. domesticus); k, ZALENDE/Ei (M. m. domesticus) (m.m.dom.); l, SF/CamEi (M. musculus); m, PERA/Rk (M. musculus); n, PERC/Ei (M. musculus) (m.mus.); o, SPRET/Ei (M. spretus) (m.spr.); p, PANCEVO/Ei (M. spicelegus); q, Halbturn (M. spicelegus) (m.spi.); r, M. cervicolor (m.cer.); s, M. caroli (m.car.); t, M. cookii (m.coo.). Some known provirus loci, Bxv1 (23), Xmv10, and Xmv28 (19), are shown by arrows. The approximate positions of molecular markers are also shown.
FIG. 5
Distribution of polytropic MLV-related proviruses in wild mice. Analysis of _Pvu_II-digested mouse DNAs was performed by hybridization of the DNA in dried agarose gels with the P-I-specific oligonucleotide probe (Pltr) (A), the P-II-specific oligonucleotide probe (Mltr) (B), the P-IV-specific oligonucleotide probe (KT-58) (C), and the P-V-specific oligonucleotide probe (KT-76) (D). (A and B) Lanes: a, C57BL/6J; b, CASA/Rk (M. m. castaneus); c, SF/CamEi (M. musculus); d, PERA/Rk (M. musculus); e, PERC/Ei (M. musculus) (m.mus.); f, SPRET/Ei (M. spretus) (m.spr.); g, PANCEVO/Ei (M. spicelegus); h, Halbturn (M. spicelegus) (m.spi.); i, M. cervicolor (m.cer.); j, M. caroli (m.car.); k, M. cookii (m.coo.). (C and D) a, CZECH II/Ei (M. m. musculus); b, ZALENDE/Ei (M. m. domesticus) (m.mus.); c, SPRET/Ei (M. spretus) (m.spr.); d, PANCEVO/Ei (M. spicelegus); e, Halbturn (M. spicelegus) (m.spi.); f, M. cervicolor (m.cer.); g, M. caroli (m.car.); h, M. cookii (m.coo.). The approximate positions of molecular markers are also shown.
FIG. 6
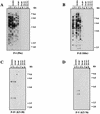
Detection of fragments of the M. spicelegus endogenous provirus (HEMV). (A) The locations of PCR primers and the oligonucleotide probes used. The approximate positions of the SU and TM regions of the env gene and the U3 and R regions of the LTR are indicated. Two hypervariable regions (VRA and VRB) in the SU region are also shown by boxes. (B) Detection of the HEMV provirus. Analysis of _Pvu_II-digested mouse DNAs was performed by using the HEMV-specific oligonucleotide probe KT-61. Lanes: a, CZECH II/Ei (M. musculus); b, SPRET/Ei (M. spretus); c, PANCEVO/Ei (M. spicelegus); d, Halbturn (M. spicelegus); e, M. cervicolor; f, M. caroli; g, M. cookii. The approximate positions of molecular markers are also shown. (C) Amino acid sequences of the VRA and VRB regions of the HEMV env gene are aligned with the analogous regions of several type C retroviruses. Abbreviations and strains of the viruses included in this alignment are as follows: P-I (MX27) (49); X-I (NZB) (41); Ampho, amphotropic MLV (4070A) (42); Eco, ecotropic MLV (Akv) (58); MDEV, M. dunni endogenous virus (62); GalV, (U20589); FeLV, feline leukemia virus subgroup A (Glasgow-1) (48).
FIG. 7
Alignment of HEMV U3 sequences. (A) The U3 region of the HEMV provirus was obtained as described in the text and is aligned with the corresponding sequences from the X-IV (Mxv11), X-I (NFS-Th-1) (28), P-I (MX27) (49), and ecotropic (Akv) (58) proviruses. Dots indicate nucleotide identity. Dashes indicate absence of a nucleotide. The sequence of an oligonucleotide probe, KT-59/60, reactive with X-IV proviruses is underlined. Direct repeats in the enhancer regions found in ecotropic proviruses are not shown. The _Pst_I recognition site is also shown. (B) Lack of the region 6 direct repeat in HEMV. The 3′ portion of the HEMV U3 sequence is aligned with the analogous regions of the several type C retroviruses. Abbreviations and strains of the viruses included in this alignment are as follows: GaLV-SE and GaLV-SF, SEATO and San Francisco isolates (56); FeLV-A and FeLV-B, feline leukemia virus, subgroup A (Glasgow-1) and subgroup B (Gardner-Arnstein) (48); X-I, NFS-Th-1 (28).
FIG. 8
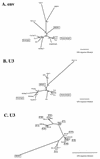
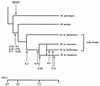
Phylogenetic analysis of the nonecotropic U3 proviruses. Unrooted phylogenetic trees for the env (A) and U3 (B and C) regions of HEMV and several type C leukemia viruses were estimated by neighbor joining. Branch lengths are drawn to scale. To illustrate consistency all bootstrap values obtained with 1,000 replications of bootstrap sampling are shown. Viruses used for this analysis are as follows: ecotropic viruses, Akv (58), Moloney (47), Friend (FE29 strain) (43), Fv4 (M33884), Cas-Br-E (P08360), and HoMuLV (60); nonecotropic viruses, P-I (MX27) (49), P-II (MX33) (49), and X-I (NFS-Th-1) (28); amphotropic viruses, (4070A) (42), MDEV (62), GaLV (SEATO) (56), and feline leukemia virus subgroup A (FeLV-A) (Glasgow-1) (48).
FIG. 9
A possible evolutionary scheme relating nonecotropic MLV U3 regions. Schematic representations of the U3 structures are shown. Locations of the UCR and CAT box are also shown. Corresponding clones are shown on the right of the structures. Clone pRFM17 was described previously (8). Note that this scheme is parsimonious with respect to insertions and deletions. Discrepancies relative to the tree shown in Fig. 8 probably stem from these events having occurred multiple times.
FIG. 10
Coevolution of MLVs and wild mice. A schematic phylogenetic relationship of wild-mouse species is shown. Possible periods of provirus integration are represented by arrows. The bar under the tree indicates the approximate time scale. MYA, million years ago.
Similar articles
- Characterization of hortulanus endogenous murine leukemia virus, an endogenous provirus that encodes an infectious murine leukemia virus of a novel subgroup.
Tipper CH, Bencsics CE, Coffin JM. Tipper CH, et al. J Virol. 2005 Jul;79(13):8316-29. doi: 10.1128/JVI.79.13.8316-8329.2005. J Virol. 2005. PMID: 15956577 Free PMC article. - Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice.
Tomonaga K, Coffin JM. Tomonaga K, et al. J Virol. 1998 Oct;72(10):8289-300. doi: 10.1128/JVI.72.10.8289-8300.1998. J Virol. 1998. PMID: 9733873 Free PMC article. - Antigenic subclasses of polytropic murine leukemia virus (MLV) isolates reflect three distinct groups of endogenous polytropic MLV-related sequences in NFS/N mice.
Evans LH, Lavignon M, Taylor M, Alamgir AS. Evans LH, et al. J Virol. 2003 Oct;77(19):10327-38. doi: 10.1128/jvi.77.19.10327-10338.2003. J Virol. 2003. PMID: 12970417 Free PMC article. - The mouse "xenotropic" gammaretroviruses and their XPR1 receptor.
Kozak CA. Kozak CA. Retrovirology. 2010 Nov 30;7:101. doi: 10.1186/1742-4690-7-101. Retrovirology. 2010. PMID: 21118532 Free PMC article. Review. - Evolution of different antiviral strategies in wild mouse populations exposed to different gammaretroviruses.
Kozak CA. Kozak CA. Curr Opin Virol. 2013 Dec;3(6):657-63. doi: 10.1016/j.coviro.2013.08.001. Epub 2013 Aug 28. Curr Opin Virol. 2013. PMID: 23992668 Free PMC article. Review.
Cited by
- ERE database: a database of genomic maps and biological properties of endogenous retroviral elements in the C57BL/6J mouse genome.
Kao D, Hsu K, Chiu S, Tu V, Chew A, Lee KH, Lee YK, Kwon DN, Greenhalgh DG, Cho K. Kao D, et al. Genomics. 2012 Sep;100(3):157-61. doi: 10.1016/j.ygeno.2012.06.002. Epub 2012 Jun 9. Genomics. 2012. PMID: 22691267 Free PMC article. - Selective up-regulation of intact, but not defective env RNAs of endogenous modified polytropic retrovirus by the Sgp3 locus of lupus-prone mice.
Yoshinobu K, Baudino L, Santiago-Raber ML, Morito N, Dunand-Sauthier I, Morley BJ, Evans LH, Izui S. Yoshinobu K, et al. J Immunol. 2009 Jun 15;182(12):8094-103. doi: 10.4049/jimmunol.0900263. J Immunol. 2009. PMID: 19494335 Free PMC article. - Sgp3 and Sgp4 control expression of distinct and restricted sets of xenotropic retroviruses encoding serum gp70 implicated in murine lupus nephritis.
Kihara M, Leroy V, Baudino L, Evans LH, Izui S. Kihara M, et al. J Autoimmun. 2011 Dec;37(4):311-8. doi: 10.1016/j.jaut.2011.09.001. Epub 2011 Oct 7. J Autoimmun. 2011. PMID: 21982749 Free PMC article. - Characterization of hortulanus endogenous murine leukemia virus, an endogenous provirus that encodes an infectious murine leukemia virus of a novel subgroup.
Tipper CH, Bencsics CE, Coffin JM. Tipper CH, et al. J Virol. 2005 Jul;79(13):8316-29. doi: 10.1128/JVI.79.13.8316-8329.2005. J Virol. 2005. PMID: 15956577 Free PMC article. - Lipopolysaccharide stress induces cell-type specific production of murine leukemia virus type-endogenous retroviral virions in primary lymphoid cells.
Kwon DN, Lee YK, Greenhalgh DG, Cho K. Kwon DN, et al. J Gen Virol. 2011 Feb;92(Pt 2):292-300. doi: 10.1099/vir.0.023416-0. Epub 2010 Oct 21. J Gen Virol. 2011. PMID: 20965985 Free PMC article.
References
- Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources