Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans - PubMed (original) (raw)
Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans
S Parisi et al. Mol Cell Biol. 1999 May.
Abstract
Our work and that of others defined mitosis-specific (Rad21 subfamily) and meiosis-specific (Rec8 subfamily) proteins involved in sister chromatid cohesion in several eukaryotes, including humans. Mutation of the fission yeast Schizosaccharomyces pombe rec8 gene was previously shown to confer a number of meiotic phenotypes, including strong reduction of recombination frequencies in the central region of chromosome III, absence of linear element polymerization, reduced pairing of homologous chromosomes, reduced sister chromatid cohesion, aberrant chromosome segregation, defects in spore formation, and reduced spore viability. Here we extend the description of recombination reduction to the central regions of chromosomes I and II. We show at the protein level that expression of rec8 is meiosis specific and that Rec8p localizes to approximately 100 foci per prophase nucleus. Rec8p was present in an unphosphorylated form early in meiotic prophase but was phosphorylated prior to meiosis I, as demonstrated by analysis of the mei4 mutant blocked before meiosis I. Evidence for the persistence of Rec8p beyond meiosis I was obtained by analysis of the mutant mes1 blocked before meiosis II. A human gene, which we designate hrec8, showed significant primary sequence similarity to rec8 and was mapped to chromosome 14. High mRNA expression of mouse and human rec8 genes was found only in germ line cells, specifically in testes and, interestingly, in spermatids. hrec8 was also expressed at a low level in the thymus. Sequence similarity and testis-specific expression indicate evolutionarily conserved functions of Rec8p in meiosis. Possible roles of Rec8p in the integration of different meiotic events are discussed.
Figures
FIG. 1
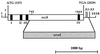
Schematic diagram of a 2.3-kb fragment carrying the rec8 gene and of the rec8::ura4 insertion mutation. The open box indicates the ORF of the rec8 gene. Solid boxes indicate the four introns at the following nucleotide positions: I at 213 to 255, II at 444 to 495, III at 593 to 644, and IV at 1895 to 1934. The putative TATA box (T), translation start and stop sites, and three potential polyadenylation signals (A1 to A3) are also indicated. In contrast to the originally published rec8 sequence (742 to 1923) (46), the actual gene structure contains four introns that were first identified on the basis of the published splice consensus sequences for S. pombe (64) (reference and data not shown). The existence of these introns was confirmed as described in Materials and Methods. The A of the ATG is position 157, and the A of the TGA is position 2029. To produce the rec8::ura4 deletion mutant, the ura4 marker gene was used to replace the fragment between the 5′ _Nsi_I and 3′ _Nhe_I restriction sites.
FIG. 2
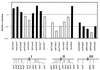
Meiotic recombinant frequencies in rec8 mutants. For each interval, the frequency is expressed relative to the one measured in the rec+ cross. The solid bars represent the data from reference , and the open bars represent those obtained from rec8::ura4 crosses. Recombination frequencies were measured twice in every interval in the rec+ and the rec8::ura4 backgrounds. The approximate positions of the genes on the chromosomes are shown below the histogram.
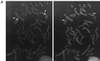
FIG. 3
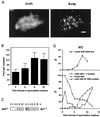
Immunolocalization of Rec8p-staining foci and rec8 mRNA expression in relation to a time course of cytological events during wild-type meiosis. Nuclear spreads were prepared from wild-type (PA39) diploid cells 0, 2, 4, 6, 8, and 10 h after transfer to meiosis medium, and all data are from the same meiosis time course experiment. (A) Example of a spread wild-type nucleus 8 h after induction of meiosis. Rec8p was visualized with an affinity purified polyclonal anti-Rec8p antibody (right), and the DNA was counterstained with DAPI (left). Bar, 2 μm. (B) Quantitation of Rec8p-staining foci during meiosis. The number of foci was determined in samples of 30 well-spread nuclei at the indicated time points. The error bars indicate 1 standard deviation. (C) Induction of rec8 mRNA expression during wild-type meiosis. Northern blots of 20 μg of total RNA at the indicated time points were hybridized with a _rec8_-specific probe (left) and later hybridized to a _byr1_-specific cDNA probe to verify loading (right). (D) Timing of cytological events during wild-type (WT) meiosis. The number of horse-tail nuclei, which are markers of meiotic prophase, was determined by DAPI staining, and the percentage of cells with 4C DNA content was measured by flow cytometry. Cells with more than one nucleus represent the percentage of the cells that have completed the first meiotic division. The increase in the number of cells containing two nuclei 1 h after induction of meiosis is due to the mitotic division of cells in G2 phase. This last mitotic division must occur before cells can enter meiosis from the G1 phase (21).
FIG. 4
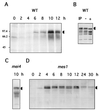
Expression and phosphorylation of Rec8p during meiosis. (A) Protein extracts were prepared at different time points of a meiosis time course of the wild-type (PA39; WT) diploid. Aliquots of 300 μg were subjected to SDS-PAGE and Western blot analysis with affinity-purified anti-Rec8p antiserum. Arrowheads indicate the two forms of Rec8p. (B) Phosphatase treatment of Rec8p from wild-type (WT) cells (strain PA39) 8 h after induction of meiosis. IP, immunoprecipitate; −, no phosphatase treatment; +, phosphatase treatment. (C) Immunoprecipitation of Rec8p from mei4 (PA41) cell extracts 10 h after a shift to sporulation medium. (D) Meiotic time course of the mes1 diploid strain PA42. Protein extracts were prepared at the indicated time points, and aliquots of 150 μg were subjected to SDS-PAGE and Western blot analysis with affinity-purified rat anti-Rec8p antiserum.
FIG. 5
rad21/rec8 gene family. (A) Alignment of the predicted amino acid sequence of S. pombe Rec8p with human Rec8p, an S. cerevisiae Rec8p homolog (Rec8psc), and the founding family member, Rad21p. Letters in black boxes represent identical amino acids in at least two species, whereas those in gray boxes represent similar (P, A, G, S, and T; E, D, N, and Q; V, I, L, and M; F, W, and Y; R, K, and H) amino acids. Gaps introduced into the sequences for alignment optimization are shown as dots. Numbers denote amino acid numbers in the sequence. (B) Alignment of the conserved N- and C-terminal amino acid regions (top and bottom, respectively) of rad21/rec8 gene family members from different species. Sequences shown here are as follows: rec8sc, S. cerevisiae Rec8p homolog (GenBank accession no. U31900); rec8 (reference and this study); hrec8 (present study); SCC1, S. cerevisiae Rad21p homolog (28, 49) (GenBank U23759); rad21, S. pombe Rad21p (8); cer21a and cer21b, first and second C. elegans Rad21p homologs (reference and this study) (GenBank U40029 and U38377, respectively); mrad21 and hrad21, mHR21sp and hHR21sp (mouse and human Rad21p homologs) (48) (GenBank X98293 and X98294, respectively); and PSEUDO, hHR21sp pseudogene on chromosome X (GenBank HSU85A3). Rec8sc, Rec8p, and hRec8p are the S. cerevisiae, S. pombe, and human Rec8 proteins that are the subject of this publication. Amino acid shading and general features of the alignment are as in panel A. Regions of less highly conserved sequence exist between the blocks and are indicated by dashes and arrowheads. The arrowhead at the end of cer21a represents 22 nonconserved amino acids at its C terminus. The arrowhead at the beginning of cer21b represents 55 nonconserved amino acids at its N terminus. (C) Unrooted phylogenetic tree of Rad21 and Rec8 proteins from different species. Evolutionary distances between the proteins from different species are indicated by the lengths of the lines in the figure. The sequences represented here are the same as in panel B, except that for reasons of clarity, the mHR21sp protein (and the pseudogene) are not on the tree because of their evolutionary proximity to hHR21spp.
FIG. 5
rad21/rec8 gene family. (A) Alignment of the predicted amino acid sequence of S. pombe Rec8p with human Rec8p, an S. cerevisiae Rec8p homolog (Rec8psc), and the founding family member, Rad21p. Letters in black boxes represent identical amino acids in at least two species, whereas those in gray boxes represent similar (P, A, G, S, and T; E, D, N, and Q; V, I, L, and M; F, W, and Y; R, K, and H) amino acids. Gaps introduced into the sequences for alignment optimization are shown as dots. Numbers denote amino acid numbers in the sequence. (B) Alignment of the conserved N- and C-terminal amino acid regions (top and bottom, respectively) of rad21/rec8 gene family members from different species. Sequences shown here are as follows: rec8sc, S. cerevisiae Rec8p homolog (GenBank accession no. U31900); rec8 (reference and this study); hrec8 (present study); SCC1, S. cerevisiae Rad21p homolog (28, 49) (GenBank U23759); rad21, S. pombe Rad21p (8); cer21a and cer21b, first and second C. elegans Rad21p homologs (reference and this study) (GenBank U40029 and U38377, respectively); mrad21 and hrad21, mHR21sp and hHR21sp (mouse and human Rad21p homologs) (48) (GenBank X98293 and X98294, respectively); and PSEUDO, hHR21sp pseudogene on chromosome X (GenBank HSU85A3). Rec8sc, Rec8p, and hRec8p are the S. cerevisiae, S. pombe, and human Rec8 proteins that are the subject of this publication. Amino acid shading and general features of the alignment are as in panel A. Regions of less highly conserved sequence exist between the blocks and are indicated by dashes and arrowheads. The arrowhead at the end of cer21a represents 22 nonconserved amino acids at its C terminus. The arrowhead at the beginning of cer21b represents 55 nonconserved amino acids at its N terminus. (C) Unrooted phylogenetic tree of Rad21 and Rec8 proteins from different species. Evolutionary distances between the proteins from different species are indicated by the lengths of the lines in the figure. The sequences represented here are the same as in panel B, except that for reasons of clarity, the mHR21sp protein (and the pseudogene) are not on the tree because of their evolutionary proximity to hHR21spp.
FIG. 5
rad21/rec8 gene family. (A) Alignment of the predicted amino acid sequence of S. pombe Rec8p with human Rec8p, an S. cerevisiae Rec8p homolog (Rec8psc), and the founding family member, Rad21p. Letters in black boxes represent identical amino acids in at least two species, whereas those in gray boxes represent similar (P, A, G, S, and T; E, D, N, and Q; V, I, L, and M; F, W, and Y; R, K, and H) amino acids. Gaps introduced into the sequences for alignment optimization are shown as dots. Numbers denote amino acid numbers in the sequence. (B) Alignment of the conserved N- and C-terminal amino acid regions (top and bottom, respectively) of rad21/rec8 gene family members from different species. Sequences shown here are as follows: rec8sc, S. cerevisiae Rec8p homolog (GenBank accession no. U31900); rec8 (reference and this study); hrec8 (present study); SCC1, S. cerevisiae Rad21p homolog (28, 49) (GenBank U23759); rad21, S. pombe Rad21p (8); cer21a and cer21b, first and second C. elegans Rad21p homologs (reference and this study) (GenBank U40029 and U38377, respectively); mrad21 and hrad21, mHR21sp and hHR21sp (mouse and human Rad21p homologs) (48) (GenBank X98293 and X98294, respectively); and PSEUDO, hHR21sp pseudogene on chromosome X (GenBank HSU85A3). Rec8sc, Rec8p, and hRec8p are the S. cerevisiae, S. pombe, and human Rec8 proteins that are the subject of this publication. Amino acid shading and general features of the alignment are as in panel A. Regions of less highly conserved sequence exist between the blocks and are indicated by dashes and arrowheads. The arrowhead at the end of cer21a represents 22 nonconserved amino acids at its C terminus. The arrowhead at the beginning of cer21b represents 55 nonconserved amino acids at its N terminus. (C) Unrooted phylogenetic tree of Rad21 and Rec8 proteins from different species. Evolutionary distances between the proteins from different species are indicated by the lengths of the lines in the figure. The sequences represented here are the same as in panel B, except that for reasons of clarity, the mHR21sp protein (and the pseudogene) are not on the tree because of their evolutionary proximity to hHR21spp.
FIG. 6
Chromosomal assignment and tissue specificity of mammalian rec8 mRNA expression. (A) FISH was used for hrec8 chromosomal assignment; biotinylated full-length hrec8 cDNA was hybridized with human metaphase spreads. (Left) Specific double-hybridization signals on chromosome 14q11.2-12 (arrowheads). (Right) DAPI staining of the same metaphase spread. (B) To examine mammalian rec8 mRNA expression, Northern blots of RNA from various mouse and human tissues and different testis fractions were hybridized with an hrec8 cDNA probe (top). (Top left) Single lane from a previously reported total RNA blot of mouse testis (Fig. 4 of reference 48); since no transcripts were detected in other tissues (thymus, brain, muscle, kidney, heart, liver, spleen, and ovary), these lanes are not shown here. (Top middle) Murine testis fractions representing meiotic (spermatocytes) and postmeiotic (a mixture of round and elongating spermatids) fractions. (Top right) Clontech multiple-human-tissue blot (no. 7754-1). There was some cross-hybridization of the hrec8 probe with trace amounts of 28S rRNA, as previously observed by us (data not shown). (Bottom left and middle) Ethidium bromide staining of rRNA indicating lane loading. (Bottom right) The multiple tissue blot was rehybridized with a β-actin control for lane loading. The positions of the 28S and 18S rRNA bands are shown on the left, while the transcript sizes (in kilobases) are indicated on the right.
FIG. 6
Chromosomal assignment and tissue specificity of mammalian rec8 mRNA expression. (A) FISH was used for hrec8 chromosomal assignment; biotinylated full-length hrec8 cDNA was hybridized with human metaphase spreads. (Left) Specific double-hybridization signals on chromosome 14q11.2-12 (arrowheads). (Right) DAPI staining of the same metaphase spread. (B) To examine mammalian rec8 mRNA expression, Northern blots of RNA from various mouse and human tissues and different testis fractions were hybridized with an hrec8 cDNA probe (top). (Top left) Single lane from a previously reported total RNA blot of mouse testis (Fig. 4 of reference 48); since no transcripts were detected in other tissues (thymus, brain, muscle, kidney, heart, liver, spleen, and ovary), these lanes are not shown here. (Top middle) Murine testis fractions representing meiotic (spermatocytes) and postmeiotic (a mixture of round and elongating spermatids) fractions. (Top right) Clontech multiple-human-tissue blot (no. 7754-1). There was some cross-hybridization of the hrec8 probe with trace amounts of 28S rRNA, as previously observed by us (data not shown). (Bottom left and middle) Ethidium bromide staining of rRNA indicating lane loading. (Bottom right) The multiple tissue blot was rehybridized with a β-actin control for lane loading. The positions of the 28S and 18S rRNA bands are shown on the left, while the transcript sizes (in kilobases) are indicated on the right.
Similar articles
- The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis.
Molnar M, Bähler J, Sipiczki M, Kohli J. Molnar M, et al. Genetics. 1995 Sep;141(1):61-73. doi: 10.1093/genetics/141.1.61. Genetics. 1995. PMID: 8536990 Free PMC article. - A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction.
Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. Pasierbek P, et al. Genes Dev. 2001 Jun 1;15(11):1349-60. doi: 10.1101/gad.192701. Genes Dev. 2001. PMID: 11390355 Free PMC article. - Meiotic chromosome dynamics dependent upon the rec8(+), rec10(+) and rec11(+) genes of the fission yeast Schizosaccharomyces pombe.
Krawchuk MD, DeVeaux LC, Wahls WP. Krawchuk MD, et al. Genetics. 1999 Sep;153(1):57-68. doi: 10.1093/genetics/153.1.57. Genetics. 1999. PMID: 10471700 Free PMC article. - Sister chromatid cohesion and recombination in meiosis.
van Heemst D, Heyting C. van Heemst D, et al. Chromosoma. 2000;109(1-2):10-26. doi: 10.1007/s004120050408. Chromosoma. 2000. PMID: 10855491 Review. - The nature of meiotic chromosome dynamics and recombination in budding yeast.
Hong S, Joo JH, Yun H, Kim K. Hong S, et al. J Microbiol. 2019 Apr;57(4):221-231. doi: 10.1007/s12275-019-8541-9. Epub 2019 Jan 22. J Microbiol. 2019. PMID: 30671743 Review.
Cited by
- The C-terminus of S. pombe DDK subunit Dfp1 is required for meiosis-specific transcription and cohesin cleavage.
Le AH, Mastro TL, Forsburg SL. Le AH, et al. Biol Open. 2013 Jun 11;2(7):728-38. doi: 10.1242/bio.20135173. Print 2013 Jul 15. Biol Open. 2013. PMID: 23862021 Free PMC article. - Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3.
Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C. Eijpe M, et al. J Cell Biol. 2003 Mar 3;160(5):657-70. doi: 10.1083/jcb.200212080. J Cell Biol. 2003. PMID: 12615909 Free PMC article. - Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination.
Grishchuk AL, Kohli J. Grishchuk AL, et al. Genetics. 2003 Nov;165(3):1031-43. doi: 10.1093/genetics/165.3.1031. Genetics. 2003. PMID: 14668362 Free PMC article. - Increased Sampling and Intracomplex Homologies Favor Vertical Over Horizontal Inheritance of the Dam1 Complex.
van Rooijen LE, Tromer EC, van Hooff JJE, Kops GJPL, Snel B. van Rooijen LE, et al. Genome Biol Evol. 2023 Mar 3;15(3):evad017. doi: 10.1093/gbe/evad017. Genome Biol Evol. 2023. PMID: 36790109 Free PMC article. - Age-Related Loss of Cohesion: Causes and Effects.
Cheng JM, Liu YX. Cheng JM, et al. Int J Mol Sci. 2017 Jul 22;18(7):1578. doi: 10.3390/ijms18071578. Int J Mol Sci. 2017. PMID: 28737671 Free PMC article. Review.
References
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Baker B S, Carpenter A T C, Esposito M S, Esposito R E, Sandler L. The genetic control of meiosis. Annu Rev Genet. 1976;10:53–134. - PubMed
- Bashkirov, V., and W.-D. Heyer. Personal communication.
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases