Activation of somatostatin receptor II expression by transcription factors MIBP1 and SEF-2 in the murine brain - PubMed (original) (raw)
Activation of somatostatin receptor II expression by transcription factors MIBP1 and SEF-2 in the murine brain
U Dörflinger et al. Mol Cell Biol. 1999 May.
Abstract
Somatostatin receptor type II expression in the mammalian brain displays a spatially and temporally very restricted pattern. In an investigation of the molecular mechanisms controlling these patterns, we have recently shown that binding of the transcription factor SEF-2 to a novel initiator element in the SSTR-2 promoter is essential for SSTR-2 gene expression. Further characterization of the promoter identified a species-conserved TC-rich enhancer element. By screening a mouse brain cDNA expression library, we cloned a cDNA encoding the transcription factor MIBP1. MIBP1 interacts specifically with both the TC box in the SSTR-2 promoter and with the SEF-2 initiator-binding protein to enhance transcription from the basal SSTR-2 promoter. We then investigated SSTR-2, SEF-2, and MIBP1 mRNA expression patterns in the developing and adult murine brain by Northern blotting and in situ hybridization. While SEF-2 is widely expressed in many neuronal and nonneuronal tissues, MIBP1 expression overlapped precisely with expression of SSTR-2 in the frontal cortex and hippocampus. In summary, our data for the first time define a regulatory role for the transcription factor MIBP1 in mediating spatially and temporally regulated SSTR-2 expression in the brain.
Figures
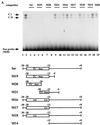
FIG. 1
(A) Nucleotide sequence of the human (hu) SSTR-2 promoter (top) from positions +8 to −35 with respect to the mRNA initiation site and comparison to the murine (mu) SSTR-2 promoter (bottom). A conserved TC-rich sequence (TC box) is printed in boldface, and the E box from residues −11 to −6 is shown in italics. (B) Identification of _cis_-regulatory elements in the human SSTR-2 promoter. The indicated reporter plasmids (500 ng) were transfected into N2A, NGP, and GHFT-1 cells (rows 1 to 10). The mRNA initiation sites are indicated by two arrows. The E box and the TC box are shown as rectangles. The numbers indicate positions of the respective nucleotides within the human SSTR-2 promoter. The reporter M6-LUC, which contains a single point mutation in the E box (MUT), is shown in row 8, and reporters which contain the double point mutations in the TC box (CC to GG) are shown in rows 5 to 7.
FIG. 2
Comparison of the murine (mu), rat, and human (hu) MIBP1 peptide sequences. The rat and human amino acid residues differing from the respective residues of the murine protein are indicated below the murine sequence. Especially well conserved motifs include the N- and C-terminal zinc fingers (doubly underlined), the putative nuclear localization signal (boxed), and the acidic region (underlined).
FIG. 2
Comparison of the murine (mu), rat, and human (hu) MIBP1 peptide sequences. The rat and human amino acid residues differing from the respective residues of the murine protein are indicated below the murine sequence. Especially well conserved motifs include the N- and C-terminal zinc fingers (doubly underlined), the putative nuclear localization signal (boxed), and the acidic region (underlined).
FIG. 3
Sequence-specific binding of MIBP1 to the human SSTR-2 promoter. (A) Electrophoretic mobility shift assays were performed with a 32P-labelled oligonucleotide containing the promoter sequence from positions −28 to −18 (M20) and in vitro-translated MIBP1 protein (lane 2). Competition experiments were performed with 50- and 100-fold molar excesses of the indicated competitors. The MIBP1 TC-box complexes CI and CII are competed by the oligonucleotides Inr, M19, and M20. Oligonucleotides M21, containing a partially deleted TC box, and M14, representing the 3′ promoter region harboring the E box, fail to compete. Competition by oligonucleotides M16, M17, and M18, representing point-mutated TC boxes, is drastically impaired. As a control an unrelated competitor, M50, also failed to compete. (B) The C-terminal MIBP1 zinc finger binds to the SSTR-2 promoter. Electrophoretic mobility shift assays were performed with the Inr oligonucleotide from nucleotides −30 to +8 and purified recombinant GST fusion protein (lane 1). A 100-fold molar excess of unlabelled Inr oligonucleotide, oligonucleotide M1, or oligonucleotide M19 competed with formation of the MIBP1-Inr complexes CI and CII. The control oligonucleotides M2 and M21 failed to compete. (C) MIBP1 binds to the SSTR-2 promoter in vivo. Whole-cell extract prepared from GHFT-1 cells (4 μl) was incubated with 1 ng of 32P-labelled oligonucleotide M19 (lanes 2 to 7). Complex CI is specifically competed by a 100-fold molar excess of oligonucleotides M19 and Inr. In contrast, the unrelated oligonucleotide M50 does not compete. To establish the composition of complex CI, the electrophoretic mobility shift assay mixture was challenged with an antiserum directed against the C-terminal zinc finger domain of MIBP1 (lane 6) or an unrelated anti-GST control antibody (lane 7). The very faint supershifted MIBP1 complex is labelled α-P-M-C.
FIG. 3
Sequence-specific binding of MIBP1 to the human SSTR-2 promoter. (A) Electrophoretic mobility shift assays were performed with a 32P-labelled oligonucleotide containing the promoter sequence from positions −28 to −18 (M20) and in vitro-translated MIBP1 protein (lane 2). Competition experiments were performed with 50- and 100-fold molar excesses of the indicated competitors. The MIBP1 TC-box complexes CI and CII are competed by the oligonucleotides Inr, M19, and M20. Oligonucleotides M21, containing a partially deleted TC box, and M14, representing the 3′ promoter region harboring the E box, fail to compete. Competition by oligonucleotides M16, M17, and M18, representing point-mutated TC boxes, is drastically impaired. As a control an unrelated competitor, M50, also failed to compete. (B) The C-terminal MIBP1 zinc finger binds to the SSTR-2 promoter. Electrophoretic mobility shift assays were performed with the Inr oligonucleotide from nucleotides −30 to +8 and purified recombinant GST fusion protein (lane 1). A 100-fold molar excess of unlabelled Inr oligonucleotide, oligonucleotide M1, or oligonucleotide M19 competed with formation of the MIBP1-Inr complexes CI and CII. The control oligonucleotides M2 and M21 failed to compete. (C) MIBP1 binds to the SSTR-2 promoter in vivo. Whole-cell extract prepared from GHFT-1 cells (4 μl) was incubated with 1 ng of 32P-labelled oligonucleotide M19 (lanes 2 to 7). Complex CI is specifically competed by a 100-fold molar excess of oligonucleotides M19 and Inr. In contrast, the unrelated oligonucleotide M50 does not compete. To establish the composition of complex CI, the electrophoretic mobility shift assay mixture was challenged with an antiserum directed against the C-terminal zinc finger domain of MIBP1 (lane 6) or an unrelated anti-GST control antibody (lane 7). The very faint supershifted MIBP1 complex is labelled α-P-M-C.
FIG. 3
Sequence-specific binding of MIBP1 to the human SSTR-2 promoter. (A) Electrophoretic mobility shift assays were performed with a 32P-labelled oligonucleotide containing the promoter sequence from positions −28 to −18 (M20) and in vitro-translated MIBP1 protein (lane 2). Competition experiments were performed with 50- and 100-fold molar excesses of the indicated competitors. The MIBP1 TC-box complexes CI and CII are competed by the oligonucleotides Inr, M19, and M20. Oligonucleotides M21, containing a partially deleted TC box, and M14, representing the 3′ promoter region harboring the E box, fail to compete. Competition by oligonucleotides M16, M17, and M18, representing point-mutated TC boxes, is drastically impaired. As a control an unrelated competitor, M50, also failed to compete. (B) The C-terminal MIBP1 zinc finger binds to the SSTR-2 promoter. Electrophoretic mobility shift assays were performed with the Inr oligonucleotide from nucleotides −30 to +8 and purified recombinant GST fusion protein (lane 1). A 100-fold molar excess of unlabelled Inr oligonucleotide, oligonucleotide M1, or oligonucleotide M19 competed with formation of the MIBP1-Inr complexes CI and CII. The control oligonucleotides M2 and M21 failed to compete. (C) MIBP1 binds to the SSTR-2 promoter in vivo. Whole-cell extract prepared from GHFT-1 cells (4 μl) was incubated with 1 ng of 32P-labelled oligonucleotide M19 (lanes 2 to 7). Complex CI is specifically competed by a 100-fold molar excess of oligonucleotides M19 and Inr. In contrast, the unrelated oligonucleotide M50 does not compete. To establish the composition of complex CI, the electrophoretic mobility shift assay mixture was challenged with an antiserum directed against the C-terminal zinc finger domain of MIBP1 (lane 6) or an unrelated anti-GST control antibody (lane 7). The very faint supershifted MIBP1 complex is labelled α-P-M-C.
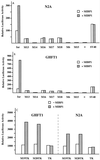
FIG. 4
(A and B) MIBP1 mediates transcriptional activation of the human SSTR-2 promoter in N2A cells (A) and GHFT-1 cells (B). The luciferase activity of the reporter constructs Inr-LUC, the mutated Inr reporter constructs M13-LUC to M18-LUC, M6-LUC, and the promoterless control plasmids basic pGL (−) or SV40-LUC is displayed as open bars. Shaded bars represent promoter activity in cells cotransfected with 200 ng of pCMX-MIBP1 expression plasmid. (C) MIBP1 mediates transcriptional activation via the TC box of the human SSTR-2 promoter. Samples (500 ng) of the reporter construct M19-TK-LUC or M20-TK-LUC containing either the Inr sequence from nucleotides −28 to −9 (TC box [Fig. 1]) or −28 to 18 in front of the thymidine kinase (TK) promoter were cotransfected with 200 ng of pCMX-MIBP1 expression plasmid (shaded bars) or with empty pCMX plasmid (open bars). As a control, the pCMX-MIBP1 expression plasmid was also cotransfected with the promoterless TK-LUC reporter.
FIG. 5
MIBP1 and SEF-2 proteins interact in vitro. The GST pull-down experiment was performed with 5 μl of in vitro-translated [35S]methionine-labelled MIBP1 protein (input) and immobilized, recombinant GST–SEF-2 fusion protein, GST–bHLH–SEF-2, GST–Δ-bHLH–SEF-2, GST-E12, or GST-E47. The control protein GST failed to interact with ivt-MIBP1.
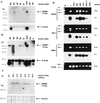
FIG. 6
Multiple tissue Northern blots of adult human (A) and adult murine (B) tissues and developmental stages E9.5 to E19.5 of murine embryogenesis (C). A single MIBP1 transcript of approximately 9.5 kb is indicated by an arrow. Abbreviations: ht (heart), br (brain), pl (placenta), lu (lung), li (liver), skm (skeletal muscle), kd (kidney), pa (pancreas), sp (spleen), te (testis). β-Actin control hybridizations or ethidium bromide-stained 18S RNA are shown below the blots. (D) Reverse transcriptase PCR amplification of SSTR-2, SEF-2, and MIBP1 transcripts with 3 μg of total cellular RNA from the neuroblastoma cell lines (N2A, NGP, and Lan-1), pituitary gland cells (GHFT-1), and epithelial cells (HeLa and COS-1). Ethidium bromide (Et-br)-stained gels and Southern blots probed with the respective phospholabelled probes are shown in parallel.
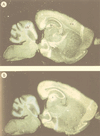
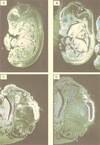
FIG. 7
Expression of SEF-2 and MIBP1 in adult mouse brain (sagittal sections) analyzed by in situ hybridization. The specificity of all the reactions was controlled by hybridizations with sense cRNA probes under identical conditions (data not shown). (A) Specific SEF-2 signals were obtained in cortex (cx), hippocampus (h), cerebellum (cb), and olfactory bulb (o). (B) Specific MIBP1 signals were obtained overlapping with all of the SEF-2-positive structures.
FIG. 8
In situ hybridization of mouse embryos of gestational stage E13.5 (A and B) and E15.5 (C and D) with SEF-2 and MIBP1 cRNA probes. The specificity of all the reactions was controlled by performing hybridization with sense probes under identical conditions (data not shown). (A and C) SEF-2 expression in the dermis (d), forebrain (f), intervertebral discs (i), gut (g), cartilage (c), and cortex (cx). (B and D) MIBP1 expression in the forebrain (primordium of the cortex [cx]), tongue (t), spinal ganglia (s), lower-jaw mesenchyme (m), and olfactory epithelium (o).
Similar articles
- The helix-loop-helix transcription factor SEF-2 regulates the activity of a novel initiator element in the promoter of the human somatostatin receptor II gene.
Pscherer A, Dörflinger U, Kirfel J, Gawlas K, Rüschoff J, Buettner R, Schüle R. Pscherer A, et al. EMBO J. 1996 Dec 2;15(23):6680-90. EMBO J. 1996. PMID: 8978694 Free PMC article. - Characterization of the biological functions of a transcription factor, c-myc intron binding protein 1 (MIBP1).
Fukuda S, Yamasaki Y, Iwaki T, Kawasaki H, Akieda S, Fukuchi N, Tahira T, Hayashi K. Fukuda S, et al. J Biochem. 2002 Mar;131(3):349-57. doi: 10.1093/oxfordjournals.jbchem.a003109. J Biochem. 2002. PMID: 11872163 - Murine helix-loop-helix transcriptional activator proteins binding to the E-box motif of the Akv murine leukemia virus enhancer identified by cDNA cloning.
Nielsen AL, Pallisgaard N, Pedersen FS, Jørgensen P. Nielsen AL, et al. Mol Cell Biol. 1992 Aug;12(8):3449-59. doi: 10.1128/mcb.12.8.3449-3459.1992. Mol Cell Biol. 1992. PMID: 1321336 Free PMC article. - Regulatory factor X4 variant 3: a transcription factor involved in brain development and disease.
Zhang D, Zeldin DC, Blackshear PJ. Zhang D, et al. J Neurosci Res. 2007 Dec;85(16):3515-22. doi: 10.1002/jnr.21356. J Neurosci Res. 2007. PMID: 17510980 Free PMC article. Review. - Transcriptional target-based expression cloning of immunoregulatory molecules.
Lamason RL, Lew SM, Pomerantz JL. Lamason RL, et al. Immunol Res. 2010 Jul;47(1-3):172-8. doi: 10.1007/s12026-009-8148-z. Immunol Res. 2010. PMID: 20069388 Free PMC article. Review.
Cited by
- FHL2, a novel tissue-specific coactivator of the androgen receptor.
Müller JM, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schüle R. Müller JM, et al. EMBO J. 2000 Feb 1;19(3):359-69. doi: 10.1093/emboj/19.3.359. EMBO J. 2000. PMID: 10654935 Free PMC article. - Novel HIVEP2 Variants in Patients with Intellectual Disability.
Park J, Colombo R, Schäferhoff K, Janiri L, Grimmel M, Sturm M, Grasshoff U, Dufke A, Haack TB, Kehrer M. Park J, et al. Mol Syndromol. 2019 Jul;10(4):195-201. doi: 10.1159/000499060. Epub 2019 Apr 3. Mol Syndromol. 2019. PMID: 31602191 Free PMC article. - Reciprocal regulation of nuclear factor kappa B and its inhibitor ZAS3 after peripheral nerve injury.
Wu LC, Goettl VM, Madiai F, Hackshaw KV, Hussain SR. Wu LC, et al. BMC Neurosci. 2006 Jan 12;7:4. doi: 10.1186/1471-2202-7-4. BMC Neurosci. 2006. PMID: 16409637 Free PMC article. - Neurodevelopmental profile of HIVEP2-related disorder.
Mo A, Snyder LG, Babington O, Chung WK, Sahin M, Srivastava S. Mo A, et al. Dev Med Child Neurol. 2022 May;64(5):654-661. doi: 10.1111/dmcn.15100. Epub 2021 Oct 26. Dev Med Child Neurol. 2022. PMID: 34704275 Free PMC article. - Transcription factor 4 and its association with psychiatric disorders.
Teixeira JR, Szeto RA, Carvalho VMA, Muotri AR, Papes F. Teixeira JR, et al. Transl Psychiatry. 2021 Jan 5;11(1):19. doi: 10.1038/s41398-020-01138-0. Transl Psychiatry. 2021. PMID: 33414364 Free PMC article. Review.
References
- Beaudet A, Greenspun D, Raelson J, Tannenbaum G S. Patterns of expression of SSTR1 and SSTR2 somatostatin receptor subtypes in the hypothalamus of the adult rat: relationship to neuroendocrine function. Neuroscience. 1995;65:551–561. - PubMed
- Beranek L, Obal F, Jr, Taishi P, Bodosi B, Laczi F, Krueger J M. Changes in rat sleep after single and repeated injections of the long-acting somatostatin analog octreotide. Am J Physiol. 1997;273:1484–1491. - PubMed
- Dimech J, Feniuk W, Latimer R D, Humphrey P P. Somatostatin-induced contraction of human isolated saphenous vein involves sst2 receptor-mediated activation of L-type calcium channels. J Cardiovasc Pharmacol. 1995;26:721–728. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases