Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae - PubMed (original) (raw)
Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae
P U Park et al. Mol Cell Biol. 1999 May.
Abstract
A cause of aging in Saccharomyces cerevisiae is the accumulation of extrachromosomal ribosomal DNA circles (ERCs). Introduction of an ERC into young mother cells shortens life span and accelerates the onset of age-associated sterility. It is important to understand the process by which ERCs are generated. Here, we demonstrate that homologous recombination is necessary for ERC formation. rad52 mutant cells, defective in DNA repair through homologous recombination, do not accumulate ERCs with age, and mutations in other genes of the RAD52 class have varying effects on ERC formation. rad52 mutation leads to a progressive delocalization of Sir3p from telomeres to other nuclear sites with age and, surprisingly, shortens life span. We speculate that spontaneous DNA damage, perhaps double-strand breaks, causes lethality in mutants of the RAD52 class and may be an initial step of aging in wild-type cells.
Figures
FIG. 1
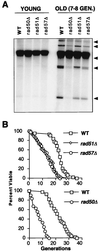
A rad52 mutant does not accumulate ERCs and has a very short life span. (A) Gel electrophoresis was performed on genomic DNA isolated from young and old wild-type (WT), sgs1, rad52, and sgs1 rad52 cells (see Materials and Methods). Various ERC species (arrowheads) similar to the ones previously observed (85) were seen in old wild-type cells and were slightly more abundant in old sgs1 cells. ERCs were undetectable in old rad52 and sgs1 rad52 cells. They were undetectable even after a longer exposure time. Average bud scar counts of old cells were as follows: wild type, 7.9 ± 1.6; sgs1, 7.6 ± 1.8; rad52, 7.4 ± 2.2; and sgs1 rad52, 7.4 ± 2.0. (B) Life span analysis was performed by standard methods as previously described (44). Average life spans were as follows: wild-type (WT), 23.5 generations; sgs1, 9.8 generations; rad52, 7.1 generations; and sgs1 rad52, 5.5 generations. (C) Homozygous rad52 diploid cells have a life span similar to that of the rad52 haploid cells. Average life spans were as follows: wild-type (WT) haploid, 24.7 generations; wild-type diploid, 23.9 generations; rad52 haploid, 7.5 generations; and rad52 diploid, 7.2 generations.
FIG. 2
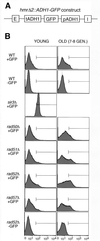
Mutants in the RAD52 epistasis group show varying degrees of premature loss of silencing at HMR. (A) A schematic diagram of the hmrΔ2::ADH1-GFP construct present in GFP-positive cells. A GFP gene driven by the constitutive ADH1 promoter was inserted between the HMR-E (E) and HMR-I (I) silencers. (B) GFP expression was efficiently silenced in a Sir-dependent manner, and silencing at HMR was lost in an age-dependent fashion. The green fluorescence level of 105 live cells was measured by FACS. The y axis indicates the number of cells, and the x axis indicates the level of green fluorescence in a log scale. Average green fluorescence intensities were as follows (average bud scar counts of old cells are given in parentheses): young wild-type (WT) +GFP, 4.65; old wild-type +GFP, 9.60 (8.9 ± 0.9); young wild-type −GFP, 2.85; old wild-type −GFP, 4.16 (8.7 ± 0.9); young sir3 +GFP, 45.16; young rad50 +GFP, 6.33; old rad50 +GFP, 16.44 (7.6 ± 1.0); young rad51 +GFP, 7.81; old rad51 +GFP, 20.30 (7.7 ± 0.8); young rad52 +GFP, 10.35; old rad52 +GFP, 26.08 (7.2 ± 0.8); young rad57 +GFP, 9.15; old rad57 +GFP, 19.56 (8.0 ± 1.0); young rad52 −GFP cells, 3.65; and old rad52 −GFP cells, 6.32 (7.5 ± 0.9).
FIG. 3
Redistribution of Sir3p from telomeres to other sites in the nucleus in old rad52 and rad50 cells. Young and old wild-type (WT), rad52, and rad50 cells were subjected to double immunolabeling with a mouse monoclonal antibody against Nop1p and affinity-purified rabbit antibodies against Sir3p (31, 46). Optical sections were acquired by charge-coupled device microscopy and the CELLscan System. The green stain represents Sir3p; the red stain represents Nop1p, a nucleolar marker; and the blue stain (DAPI) represents nuclei. In all young cells, Sir3p staining displayed perinuclear foci, indicative of telomeric localization. In old wild-type cells, Sir3p staining was observed in the nucleolus as previously observed (46). In old rad52 and rad50 cells, Sir3p staining showed a diffuse, nuclear pattern, most evident in the absence of DAPI staining. Average bud scar counts of old cells were as follows: wild type, 17.9 ± 1.3; rad50, 7.9 ± 0.8; and rad52, 7.9 ± 1.5.
FIG. 4
Role of other members of the RAD52 epistasis group in ERC formation and life span. (A) Old rad50, rad51, and rad57 cells accumulated different levels of ERCs (arrowheads) that are lower than those of the age-matched wild-type (WT) cells. Average bud scar counts of old cells were as follows: wild type, 8.2 ± 1.0; rad50, 7.9 ± 0.8; rad51, 7.8 ± 0.8; and rad57, 7.8 ± 0.8. (B) rad51 and rad57 mutants had similar life spans, which were shorter than that of wild type (WT) but longer than that of the rad52 mutant. Average life spans were as follows: wild type, 22.0 generations; rad51, 13.0 generations; and rad57, 12.5 generations. The rad50 mutant had a very short life span, similar to that of the rad52 mutant. Average life spans were as follows: wild type, 22.3 generations, and rad50, 7.3 generations.
FIG. 5
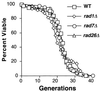
SSA, nucleotide excision, and transcription-coupled repair are not necessary for wild-type (WT) life span. Neither the rad1, the rad7, nor the rad26 mutation had a significant effect on life span. Average life spans were as follows: wild type, 22.0 generations; rad1, 20.9 generations; rad7, 22.1 generations; and rad26, 21.1 generations.
FIG. 6
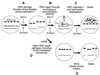
Model of yeast aging in the presence and the absence of DNA repair through homologous recombination. (A) As a young haploid cell divides, spontaneous DNA damage events, such as DSBs, occur throughout the genome including rDNA, most likely during DNA replication (59, 80, 100). (B) DSBs can efficiently be repaired through homologous recombination. DSBs that occur in the S and G2 phases of the cell cycle can be repaired with sister chromatids through interchromosomal gene conversion, by a mechanism similar to the model proposed by Szostak et al. (91). In repeated loci, including rDNA, SSA and intrachromosomal recombination can also be used for repair. The repair event at rDNA occurring through intrachromosomal recombination, if associated with a reciprocal crossover, forms an ERC. (C) As previously proposed (85), the excised ERC propagates in the mother cell with age through replication and asymmetric segregation, eventually leading to nucleolar fragmentation and death. (D) In the rad52 mutant, other DNA repair pathways, including Ku-mediated illegitimate recombination and _RAD52_-independent SSA, may try to compensate for the absence of homologous recombination and repair the DSBs. Movement of Sir proteins from telomeres to other sites in the nucleus might be linked to their involvement in such repair processes. (E) When these other repair pathways are overwhelmed, rad52 cells die due to multiple DSBs.
Similar articles
- hpr1Delta affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae.
Merker RJ, Klein HL. Merker RJ, et al. Mol Cell Biol. 2002 Jan;22(2):421-9. doi: 10.1128/MCB.22.2.421-429.2002. Mol Cell Biol. 2002. PMID: 11756539 Free PMC article. - The effects of RAD52 epistasis group genes on various types of spontaneous mitotic recombination in Saccharomyces cerevisiae.
You JC. You JC. Biochem Biophys Res Commun. 2000 Apr 2;270(1):112-8. doi: 10.1006/bbrc.2000.2382. Biochem Biophys Res Commun. 2000. PMID: 10733913 - A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52.
Firmenich AA, Elias-Arnanz M, Berg P. Firmenich AA, et al. Mol Cell Biol. 1995 Mar;15(3):1620-31. doi: 10.1128/MCB.15.3.1620. Mol Cell Biol. 1995. PMID: 7862153 Free PMC article. - The RAD52 epistasis group in mammalian double strand break repair.
Petrini JH, Bressan DA, Yao MS. Petrini JH, et al. Semin Immunol. 1997 Jun;9(3):181-8. doi: 10.1006/smim.1997.0067. Semin Immunol. 1997. PMID: 9200329 Review. - Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair.
Symington LS. Symington LS. Microbiol Mol Biol Rev. 2002 Dec;66(4):630-70, table of contents. doi: 10.1128/MMBR.66.4.630-670.2002. Microbiol Mol Biol Rev. 2002. PMID: 12456786 Free PMC article. Review.
Cited by
- SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging.
Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. Oberdoerffer P, et al. Cell. 2008 Nov 28;135(5):907-18. doi: 10.1016/j.cell.2008.10.025. Cell. 2008. PMID: 19041753 Free PMC article. - Inactivation of RAD52 and HDF1 DNA repair genes leads to premature chronological aging and cellular instability.
Mercado-Saenz S, Lopez-Diaz B, Sendra-Portero F, Martinez-Morillo M, Ruiz-Gomez MJ. Mercado-Saenz S, et al. J Biosci. 2017 Jun;42(2):219-230. doi: 10.1007/s12038-017-9684-7. J Biosci. 2017. PMID: 28569246 - Evidence that the S.cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence.
Azam M, Lee JY, Abraham V, Chanoux R, Schoenly KA, Johnson FB. Azam M, et al. Nucleic Acids Res. 2006 Jan 20;34(2):506-16. doi: 10.1093/nar/gkj452. Print 2006. Nucleic Acids Res. 2006. PMID: 16428246 Free PMC article. - Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance.
Tomaska L, Nosek J, Makhov AM, Pastorakova A, Griffith JD. Tomaska L, et al. Nucleic Acids Res. 2000 Nov 15;28(22):4479-87. doi: 10.1093/nar/28.22.4479. Nucleic Acids Res. 2000. PMID: 11071936 Free PMC article. - Genomic Copy-Number Loss Is Rescued by Self-Limiting Production of DNA Circles.
Mansisidor A, Molinar T Jr, Srivastava P, Dartis DD, Pino Delgado A, Blitzblau HG, Klein H, Hochwagen A. Mansisidor A, et al. Mol Cell. 2018 Nov 1;72(3):583-593.e4. doi: 10.1016/j.molcel.2018.08.036. Epub 2018 Oct 4. Mol Cell. 2018. PMID: 30293780 Free PMC article.
References
- Aboussekhra A, Wood R D. Repair of UV-damaged DNA by mammalian cells and Saccharomyces cerevisiae. Curr Opin Genet Dev. 1994;4:212–220. - PubMed
- Aguilera A. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr Genet. 1995;27:298–305. - PubMed
- Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials