RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains - PubMed (original) (raw)
RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains
C E Pritchard et al. J Cell Biol. 1999.
Abstract
Gle2p is implicated in nuclear export of poly(A)+ RNA and nuclear pore complex (NPC) structure and distribution in Saccharomyces cerevisiae. Gle2p is anchored at the nuclear envelope (NE) via a short Gle2p-binding motif within Nup116p called GLEBS. The molecular mechanism by which Gle2p and the Gle2p-Nup116p interaction function in mRNA export is unknown. Here we show that RAE1, the mammalian homologue of Gle2p, binds to a GLEBS-like NUP98 motif at the NPC through multiple domains that include WD-repeats and a COOH-terminal non-WD-repeat extension. This interaction is direct, as evidenced by in vitro binding studies and chemical cross-linking. Microinjection experiments performed in Xenopus laevis oocytes demonstrate that RAE1 shuttles between the nucleus and the cytoplasm and is exported from the nucleus in a temperature-dependent and RanGTP-independent manner. Docking of RAE1 to the NE is highly dependent on new mRNA synthesis. Overexpression of the GLEBS-like motif also inhibits NE binding of RAE1 and induces nuclear accumulation of poly(A)+ RNA. Both effects are abrogated either by the introduction of point mutations in the GLEBS-like motif or by overexpression of RAE1, indicating a direct role for RAE1 and the NUP98-RAE1 interaction in mRNA export. Together, our data suggest that RAE1 is a shuttling transport factor that directly contributes to nuclear export of mRNAs through its ability to anchor to a specific NUP98 motif at the NPC.
Figures
Figure 1
A GLEBS-like motif within NUP98 is necessary and sufficient for interaction with human RAE1. (A) Schematic of the NUP98 structure. Vertical bars indicate FG (phenylalanine-glycine) repeats; HA1, hemagglutinin tag; NRM, nucleoporin RNA-binding motif (shaded box); Nup116p homology region (gray box). (B) [35S]-methionine–labeled proteins immunoprecipitated with monoclonal antibody 12CA5 from HtTA cells transiently expressing an HA1-tagged version of NUP98, separated by SDS-PAGE (8% polyacrylamide) and visualized by autoradiography. A molecular weight standard is indicated at right. (C) Western blot analysis (8% polyacrylamide) of the 40-kD protein coimmunoprecipitated with HA1-NUP98 transiently expressed in HtTA cells (lanes 2 and 4). Nontransfected cells served as a negative control (lanes 1 and 3). The blots were first incubated with 12CA5 antibody (lanes 1 and 2), and then with affinity-purified anti–mouse RAE1 antibodies (lanes 3 and 4). The position of human RAE1 is indicated with an arrow. Molecular weight standards are indicated at left. (D) Western blot analysis (8% polyacrylamide) of proteins coimmunoprecipitated from HtTA lysates with anti–RAE1 (left) or anti–NUP98 antibodies (right). The antibodies used to visualize NUP98 or RAE1 proteins are indicated above the lanes. Molecular weight standards are indicated to the left. (E) Structure of NUP98 mutants used to define the GLEBS-like motif. The various NUP98 motifs are as indicated in A. (F) Western blot analysis (7% polyacrylamide) of proteins precipitated with 12CA5 antibody from HtTA cells transiently transfected with HA1-NUP98 or HA1-NUP98Δ(192–221). Molecular weight standards are indicated at left. (G) As in F for a set of HA1-tagged GLEBS-like motif mutants. Immunoprecipitated proteins were split in two equal portions, half was run through a 15% polyacrylamide gel to verify proper expression of HA1-tagged mutant peptides (top), and half on an 8% polyacrylamide gel to determine coimmunoprecipitation of RAE1. Because HA1-NUP98(150–186) protein (lane 3, top) was expressed at a lower level than the other HA1-tagged mutants, we also collected longer exposures of the RAE1 immunoblot (bottom); however, we were still unable to detect a RAE1-specific signal in lane 3. With the transfection protocol applied, levels of HA1-NUP98(150–224) were consistently higher than those of HA1-NUP98(181–224), which causes the difference in intensity of RAE1 signals in lanes 2 and 4. Molecular weight standards for each gel are indicated at left.
Figure 1
A GLEBS-like motif within NUP98 is necessary and sufficient for interaction with human RAE1. (A) Schematic of the NUP98 structure. Vertical bars indicate FG (phenylalanine-glycine) repeats; HA1, hemagglutinin tag; NRM, nucleoporin RNA-binding motif (shaded box); Nup116p homology region (gray box). (B) [35S]-methionine–labeled proteins immunoprecipitated with monoclonal antibody 12CA5 from HtTA cells transiently expressing an HA1-tagged version of NUP98, separated by SDS-PAGE (8% polyacrylamide) and visualized by autoradiography. A molecular weight standard is indicated at right. (C) Western blot analysis (8% polyacrylamide) of the 40-kD protein coimmunoprecipitated with HA1-NUP98 transiently expressed in HtTA cells (lanes 2 and 4). Nontransfected cells served as a negative control (lanes 1 and 3). The blots were first incubated with 12CA5 antibody (lanes 1 and 2), and then with affinity-purified anti–mouse RAE1 antibodies (lanes 3 and 4). The position of human RAE1 is indicated with an arrow. Molecular weight standards are indicated at left. (D) Western blot analysis (8% polyacrylamide) of proteins coimmunoprecipitated from HtTA lysates with anti–RAE1 (left) or anti–NUP98 antibodies (right). The antibodies used to visualize NUP98 or RAE1 proteins are indicated above the lanes. Molecular weight standards are indicated to the left. (E) Structure of NUP98 mutants used to define the GLEBS-like motif. The various NUP98 motifs are as indicated in A. (F) Western blot analysis (7% polyacrylamide) of proteins precipitated with 12CA5 antibody from HtTA cells transiently transfected with HA1-NUP98 or HA1-NUP98Δ(192–221). Molecular weight standards are indicated at left. (G) As in F for a set of HA1-tagged GLEBS-like motif mutants. Immunoprecipitated proteins were split in two equal portions, half was run through a 15% polyacrylamide gel to verify proper expression of HA1-tagged mutant peptides (top), and half on an 8% polyacrylamide gel to determine coimmunoprecipitation of RAE1. Because HA1-NUP98(150–186) protein (lane 3, top) was expressed at a lower level than the other HA1-tagged mutants, we also collected longer exposures of the RAE1 immunoblot (bottom); however, we were still unable to detect a RAE1-specific signal in lane 3. With the transfection protocol applied, levels of HA1-NUP98(150–224) were consistently higher than those of HA1-NUP98(181–224), which causes the difference in intensity of RAE1 signals in lanes 2 and 4. Molecular weight standards for each gel are indicated at left.
Figure 2
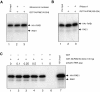
Chemical cross-linking of in vitro–translated HA1-RAE1 to E. _coli_–purified GLEBS-like motifs. (A) Purified recombinant HIS-NUP98(150–224) (∼11-kD) protein separated by SDS-PAGE (15% polyacrylamide gel) and detected by Coomassie brilliant blue (CBB; Bio-Rad Laboratories) staining. (B) Purified recombinant GST (29 kD) and GST-NUP98(150–224) (∼38 kD) protein run on a 10% polyacrylamide gel and stained with CBB. (C) Pull-down assays performed with [35S]-methionine–labeled HA1-RAE1 synthesized in vitro (45 kD), and GST- or HIS-NUP98(150–224) fusion proteins purified from E. coli. 5% input shows 5% of the labeled HA1-RAE1 protein used in each pull-down assay. Typically, the in vitro–translated RAE1 appears as a doublet, representing fragments with and without a HA1 tag (the RAE1 cDNA was cloned in pSP73 as a HA1 fusion gene that retained the endogenous RAE1 translation initiation codon). GST beads and Ni-NTA agarose acted as negative control for binding in GST-NUP98(150–224) and HIS-NUP98(150–224) pull-down assays, respectively. Comparable amounts of GST, GST-NUP98(150–224), and HIS-NUP98(150–224) proteins were used in each pull-down assay. The experiment shown is representative for two independent experiments. A cross-linked GST-NUP98(150–224)/RAE1 product of ∼84 kD and a cross-linked HIS-NUP98(150–224)/RAE1 product of ∼56 kD were obtained specifically in DSS-treated samples. Note that cross-linking of RAE1 to HIS-NUP98(150–224) was more efficient than to GST-NUP98(150–224). Molecular weight standards are indicated at right.
Figure 3
Poly(A)+ RNA is not a cofactor in the binding reaction between RAE1 and the GLEBS-like motif of NUP98 in vitro. (A) Pull-down assay performed with GST-NUP98(150– 224) fusion protein purified from E. coli and [35S]-methionine– labeled in vitro–translated HA1-RAE1 pretreated with or without Micrococcal nuclease. GST alone did not pull-down HA1-RAE1 (not shown). (B) As in A, but pretreated with or without RNase A. GST alone did not pull-down HA1-RAE1 (not shown). (C) Pull-down assay with GST-NUP98(150–224) and in vitro–translated HA1-RAE1 in the presence of various concentrations of poly(A)+ RNA isolated from HtTA cells. GST alone does not pull-down HA1-RAE1, as indicated in lane 6.
Figure 4
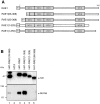
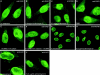
Multiple domains of RAE1 are necessary but not sufficient for interaction with NUP98. (A) Schematic representation of HA1-RAE1 deletion mutants. The four WD-repeat motifs are indicated as gray boxes. All mutants expressed an NH2-terminal HA1 tag. (B) Western blot analysis of proteins precipitated with 12CA5 antibody from lysates of HtTA cells transiently transfected with HA1-RAE1 or various deletion mutants. Precipitated proteins were visualized with 12CA5 (top) and anti–NUP98 antibodies (bottom). The HA1-RAE1 mutants used are indicated above the lanes. Molecular weight standards are indicated to the left. Results shown are representative for three independent experiments. (C–G) Confocal images detailing the subcellular distribution HA1-RAE1 deletion mutants moderately expressed in HtTA cells and stained with 12CA5 antibody. Note that the ability to coprecipitate NUP98 correlates with prominent NE staining.
Figure 4
Multiple domains of RAE1 are necessary but not sufficient for interaction with NUP98. (A) Schematic representation of HA1-RAE1 deletion mutants. The four WD-repeat motifs are indicated as gray boxes. All mutants expressed an NH2-terminal HA1 tag. (B) Western blot analysis of proteins precipitated with 12CA5 antibody from lysates of HtTA cells transiently transfected with HA1-RAE1 or various deletion mutants. Precipitated proteins were visualized with 12CA5 (top) and anti–NUP98 antibodies (bottom). The HA1-RAE1 mutants used are indicated above the lanes. Molecular weight standards are indicated to the left. Results shown are representative for three independent experiments. (C–G) Confocal images detailing the subcellular distribution HA1-RAE1 deletion mutants moderately expressed in HtTA cells and stained with 12CA5 antibody. Note that the ability to coprecipitate NUP98 correlates with prominent NE staining.
Figure 6
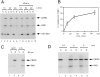
RAE1 shuttles between the nucleus and the cytoplasm. (A) A mixture of in vitro–translated [35S]-methionine–labeled CBP80, HA1-RAE1 (seen as a doublet that represents HA1-RAE1 and RAE1), and GTS-NES was injected into Xenopus laevis oocytes nuclei, either in the absence (lanes 1–6) or presence (lanes 7–12) of 80 μM Rna1p. After injection, oocytes were incubated at 20°C and protein samples from nuclear (N) and cytoplasmic (C) fractions were collected at 10, 30, or 90 min. Proteins were separated by SDS-PAGE and detected by fluorography. (B) Kinetics of RAE1 distribution after nuclear injection. Values were obtained from three experiments; error bars represent standard deviations. (C) Comparison of nuclear export of RAE1 at 20° (lanes 1 and 2) and 0°C (lanes 3 and 4). (D) Nuclear uptake of RAE1 after cytoplasmic injection after 0.2 (lanes 1 and 2), 6 (lanes 3 and 4), or 9 h (lanes 5 and 6).
Figure 5
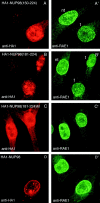
A single point mutation in a highly conserved WD-repeat residue abrogates RAE1–NUP98 interaction at the NPC. (A) A scale drawing of HA1-RAE1 depicting the positions of the highly conserved D residues within the last three WD-repeats that we individually mutated to A. (B) Schematic representation of a blade from a WD-repeat propeller structure (Garcia-Higuera et al., 1998). (a–d) The four β strands within the blade are indicated. Note that the position of the conserved D residue in the hairpin turn between b and c is highlighted (star). (C) Western blot analysis of proteins precipitated with 12CA5 antibody from lysates of HtTA cells transiently transfected with HA1-RAE1 or the point mutants. Precipitated proteins were visualized with 12CA5 (top) and anti–NUP98 antibodies (bottom). The HA1-RAE1 mutants used are indicated above the lanes. Molecular weight standards are indicated at left. Results shown are representative for three independent experiments. (D–G) Representative confocal images detailing the subcellular distribution of HA1-RAE1 mutants in HtTA cells (12CA5 antibody staining).
Figure 5
A single point mutation in a highly conserved WD-repeat residue abrogates RAE1–NUP98 interaction at the NPC. (A) A scale drawing of HA1-RAE1 depicting the positions of the highly conserved D residues within the last three WD-repeats that we individually mutated to A. (B) Schematic representation of a blade from a WD-repeat propeller structure (Garcia-Higuera et al., 1998). (a–d) The four β strands within the blade are indicated. Note that the position of the conserved D residue in the hairpin turn between b and c is highlighted (star). (C) Western blot analysis of proteins precipitated with 12CA5 antibody from lysates of HtTA cells transiently transfected with HA1-RAE1 or the point mutants. Precipitated proteins were visualized with 12CA5 (top) and anti–NUP98 antibodies (bottom). The HA1-RAE1 mutants used are indicated above the lanes. Molecular weight standards are indicated at left. Results shown are representative for three independent experiments. (D–G) Representative confocal images detailing the subcellular distribution of HA1-RAE1 mutants in HtTA cells (12CA5 antibody staining).
Figure 7
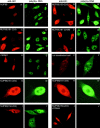
RAE1's association with the NE requires RNA polymerase II activity. (A–H) Images of HtTA cells stained for RAE1 or NUP98 after treatment with RNA polymerase inhibitors. RAE1-stained cells treated with (A) 0 μg/ml AMD for 1 h, (B) 0.04 μg/ml AMD for 1 h, (C) 5.0 μg/ml AMD for 1 h, (D) 50 μg/ml DRB for 1 h, (E) 50 μg/ml DRB for 1 h and cultured 6 h in the absence of DRB, and (F) 50 μg/ml DRB for 1 h, cultured 6 h without DRB, and finally treated again with 50 μg/ml DRB for 1 h. Images shown are representative for results obtained from three independent experiments. (G and H) Respective nontreated and 5.0 μg/ml AMD–treated HtTA cells stained with NUP98-specific antibodies. Note that NUP98 levels at the NE are not dependent on RNA polymerase II activity. (I and J) Effect of AMD treatment on HtTA cells that moderately express HA1-RAE1.
Figure 8
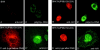
Overexpression of the GLEBS-like motif of NUP98 results in decreased levels of RAE1 at the NE. (A–D′ inclusive) Paired confocal images from HtTA cells that transiently express various forms of the GLEBS-like motif of NUP98. Cells were double-stained for HA1-tagged protein using the 12CA5 monoclonal antibody (left) and for RAE1 using affinity-purified RAE1 polyclonal rabbit antibodies. Cells shown in the confocal images are representative for results obtained in three to four independent experiments. High magnification images are given to illustrate detail. (A, A′, B, and B′) Overexpression of the GLEBS-like motif leads to a subtle but significant decrease in NE localization of RAE1. Compare NE staining of cells that do not overexpress the RAE1-binding (nt) and cells that do (t). (C, C′, D, and D′) Cells expressing a GLEBS-like motif mutant that is unable to interact with RAE1 or full-length NUP98 (D and D′) display an RAE1-distribution pattern comparable with that of wild-type HtTA cells.
Figure 9
Overexpression of the GLEBS-like motif of NUP98 results in accumulation of poly(A)+ RNA in the nucleus. (A–J′, inclusive) Paired confocal images from HtTA cells that are transiently expressing HA1-NUP98 mutants. These cells were double stained for HA1-tagged protein by immunohistochemistry using the 12CA5 monoclonal antibody (left) and for poly(A)+ RNA by in situ hybridization using a FITC-labeled oligo-(dT) 50 mer (right). The identity of the HA1-tagged NUP98 mutants is indicated as the left of each pair. The poly(A)+ RNA accumulation in B′ appears more robust than that in C′, which represents a photographic rather than a real difference. The arrow in F′ points to one of the sites of preferred poly(A)+ RNA localization (also reported by Huang et al., 1994). K and L Show poly(A)+ distribution in HtTA cells overexpressing mouse RAE1 protein and, at the same time, HA1-NUP98(150–224) or HA1-NUP98(181–224). Each row of three images shows the same field of cells stained for either HA1-tagged protein (left), ectopically expressed mouse RAE1 and native human RAE1 (middle), and poly(A)+ RNA (right). Note that mouse RAE1 overexpression restores proper mRNA export in cells expressing various forms of the GLEBS-like motif. The combined anti–HA1/poly(A)+ staining procedure does not allow extensive blocking of nonspecific 12CA5 antibody binding. Therefore, nontransfected cells display higher levels of background staining than those shown in Fig. 7.
Figure 9
Overexpression of the GLEBS-like motif of NUP98 results in accumulation of poly(A)+ RNA in the nucleus. (A–J′, inclusive) Paired confocal images from HtTA cells that are transiently expressing HA1-NUP98 mutants. These cells were double stained for HA1-tagged protein by immunohistochemistry using the 12CA5 monoclonal antibody (left) and for poly(A)+ RNA by in situ hybridization using a FITC-labeled oligo-(dT) 50 mer (right). The identity of the HA1-tagged NUP98 mutants is indicated as the left of each pair. The poly(A)+ RNA accumulation in B′ appears more robust than that in C′, which represents a photographic rather than a real difference. The arrow in F′ points to one of the sites of preferred poly(A)+ RNA localization (also reported by Huang et al., 1994). K and L Show poly(A)+ distribution in HtTA cells overexpressing mouse RAE1 protein and, at the same time, HA1-NUP98(150–224) or HA1-NUP98(181–224). Each row of three images shows the same field of cells stained for either HA1-tagged protein (left), ectopically expressed mouse RAE1 and native human RAE1 (middle), and poly(A)+ RNA (right). Note that mouse RAE1 overexpression restores proper mRNA export in cells expressing various forms of the GLEBS-like motif. The combined anti–HA1/poly(A)+ staining procedure does not allow extensive blocking of nonspecific 12CA5 antibody binding. Therefore, nontransfected cells display higher levels of background staining than those shown in Fig. 7.
Figure 10
Overexpression of the GLEBS-like motif of NUP98 does not affect nuclear protein import. (A and B) Representative images of normal and HA1-NUP98(150–224)–expressing BHK cells stained with monoclonal antibody 12CA5 and the FITC-labeled oligo-(dT) 50 mer. Note that HA1-NUP98(150–224)–expressing BHK cells accumulate poly(A)+ RNA in their nuclei. (C and D) Immunofluorescence detection of the glucocorticoid receptor–β-galactosidase fusion protein in a representative BHKgrβ cell expressing HA1-NUP98(150–224) before and 30 min after the addition of 10 mg/ml dexamethasone (DMS).
Similar articles
- Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p.
Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. Bailer SM, et al. EMBO J. 1998 Feb 16;17(4):1107-19. doi: 10.1093/emboj/17.4.1107. EMBO J. 1998. PMID: 9463388 Free PMC article. - The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins.
Wang X, Babu JR, Harden JM, Jablonski SA, Gazi MH, Lingle WL, de Groen PC, Yen TJ, van Deursen JM. Wang X, et al. J Biol Chem. 2001 Jul 13;276(28):26559-67. doi: 10.1074/jbc.M101083200. Epub 2001 May 14. J Biol Chem. 2001. PMID: 11352911 - GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function.
Murphy R, Watkins JL, Wente SR. Murphy R, et al. Mol Biol Cell. 1996 Dec;7(12):1921-37. doi: 10.1091/mbc.7.12.1921. Mol Biol Cell. 1996. PMID: 8970155 Free PMC article. - The nuclear pore complex.
Davis LI. Davis LI. Annu Rev Biochem. 1995;64:865-96. doi: 10.1146/annurev.bi.64.070195.004245. Annu Rev Biochem. 1995. PMID: 7574503 Review. - [Nuclear transport of hnRNP and mRNA].
Taura T, Siomi H, Siomi MC. Taura T, et al. Tanpakushitsu Kakusan Koso. 2000 Oct;45(14):2378-87. Tanpakushitsu Kakusan Koso. 2000. PMID: 11051839 Review. Japanese. No abstract available.
Cited by
- Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice.
Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Baker DJ, et al. J Cell Biol. 2006 Feb 13;172(4):529-40. doi: 10.1083/jcb.200507081. J Cell Biol. 2006. PMID: 16476774 Free PMC article. - Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility.
Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Griffis ER, et al. Mol Biol Cell. 2004 Apr;15(4):1991-2002. doi: 10.1091/mbc.e03-10-0743. Epub 2004 Jan 12. Mol Biol Cell. 2004. PMID: 14718558 Free PMC article. - RAE1 mediated ZEB1 expression promotes epithelial-mesenchymal transition in breast cancer.
Oh JH, Lee JY, Yu S, Cho Y, Hur S, Nam KT, Kim MH. Oh JH, et al. Sci Rep. 2019 Feb 27;9(1):2977. doi: 10.1038/s41598-019-39574-8. Sci Rep. 2019. PMID: 30814639 Free PMC article. - The mitotic checkpoint regulator RAE1 induces aggressive breast cancer cell phenotypes by mediating epithelial-mesenchymal transition.
Oh JH, Hur H, Lee JY, Kim Y, Seo Y, Kim MH. Oh JH, et al. Sci Rep. 2017 Feb 9;7:42256. doi: 10.1038/srep42256. Sci Rep. 2017. PMID: 28181567 Free PMC article. - HOXA repression is mediated by nucleoporin Nup93 assisted by its interactors Nup188 and Nup205.
Labade AS, Karmodiya K, Sengupta K. Labade AS, et al. Epigenetics Chromatin. 2016 Dec 3;9:54. doi: 10.1186/s13072-016-0106-0. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27980680 Free PMC article.
References
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiaerequired for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. - PubMed
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. - PubMed
- Audigier Y. Assays for studying functional properties of in vitro translated Gs alpha subunit. Methods Enzymol. 1994;237:239–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases