Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA - PubMed (original) (raw)
Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA
S Meeusen et al. J Cell Biol. 1999.
Abstract
Maintenance of mitochondrial DNA (mtDNA) during cell division is required for progeny to be respiratory competent. Maintenance involves the replication, repair, assembly, segregation, and partitioning of the mitochondrial nucleoid. MGM101 has been identified as a gene essential for mtDNA maintenance in S. cerevisiae, but its role is unknown. Using liquid chromatography coupled with tandem mass spectrometry, we identified Mgm101p as a component of highly enriched nucleoids, suggesting that it plays a nucleoid-specific role in maintenance. Subcellular fractionation, indirect immunofluorescence and GFP tagging show that Mgm101p is exclusively associated with the mitochondrial nucleoid structure in cells. Furthermore, DNA affinity chromatography of nucleoid extracts indicates that Mgm101p binds to DNA, suggesting that its nucleoid localization is in part due to this activity. Phenotypic analysis of cells containing a temperature sensitive mgm101 allele suggests that Mgm101p is not involved in mtDNA packaging, segregation, partitioning or required for ongoing mtDNA replication. We examined Mgm101p's role in mtDNA repair. As compared with wild-type cells, mgm101 cells were more sensitive to mtDNA damage induced by UV irradiation and were hypersensitive to mtDNA damage induced by gamma rays and H2O2 treatment. Thus, we propose that Mgm101p performs an essential function in the repair of oxidatively damaged mtDNA that is required for the maintenance of the mitochondrial genome.
Figures
Figure 1
Mgm101p is conserved. The two known Mgm101p homologues from K. lactis and S. Pombe were identified using the BLAST search algorithm (Altschul et al., 1997). Alignments were obtained using the MultAlin program (Corpet, 1988). Amino acid identities between S. cerevisae, K. lactis, and S. pombe proteins are indicated by capital letters and identity between any two is indicated by small case letters. Amino acid similarities are indicated by the following symbols: ! is anyone of IV, $ is anyone of LM, % is anyone of FY, and # is anyone of NDQEBZ. Bold overlines indicate peptides identified by liquid chromatography tandem mass-spectrometric analysis of trypsin-digested nucleoid proteins (see Materials and Methods). The asterisk and arrowhead mark the position of the mutation found in mgm101-2 (see Results) and the position of the marker insertion in strain CS6-1D (Chen et al., 1993), respectively.
Figure 2
Respiratory function is lost in mgm101-2 cells under nonpermissive conditions, but mitochondrial morphology is unaffected. W303 and JNY131(mgm101-2) cells were grown to log phase in YPG, cells were harvested, split, and washed into YPD medium at 24 or 37°C. Parallel cultures of JNY131(mgm101-2) and W303 were grown in log phase in YPD at 24 and 37°C and at fixed time points, samples were collected and analyzed for respiratory competence, and for the presence for mtDNA by DAPI staining of fixed cells (A, at indicated times) and by indirect immunofluorescence using anti-Por1p (B, 12 h) as described in Materials and Methods.
Figure 2
Respiratory function is lost in mgm101-2 cells under nonpermissive conditions, but mitochondrial morphology is unaffected. W303 and JNY131(mgm101-2) cells were grown to log phase in YPG, cells were harvested, split, and washed into YPD medium at 24 or 37°C. Parallel cultures of JNY131(mgm101-2) and W303 were grown in log phase in YPD at 24 and 37°C and at fixed time points, samples were collected and analyzed for respiratory competence, and for the presence for mtDNA by DAPI staining of fixed cells (A, at indicated times) and by indirect immunofluorescence using anti-Por1p (B, 12 h) as described in Materials and Methods.
Figure 3
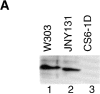
Mgm101p cofractionates with mitochondrial nucleoids. (A) Cultures of strain W303(pep4) and CS6-1D were grown in YPD. Total cell extract was prepared by lysing cells using the rapid boiling method (see Materials and Methods). A constant amount of total cell extract (0.2 OD600 units) from W303(pep4) (lane 1) and CS6-1D (lane 2) was analyzed by SDS-PAGE and immunoblotted with anti-Mgm101p antibody. (B) Cultures of strain W303(pep4) were grown in YPG. Cells were spheroplasted, lysed and fractionated as described in Materials and Methods. A constant amount (50 μg) of the total cell extract (T, lane 1), the 3,000 g supernate (LS, lane 2) and pellet fractions (LP, lane 3) and the 9,000 g supernate (lane 4) and pellet fractions (M, lane 5) was analyzed by SDS-PAGE and immunoblotted with anti-Por1p, anti-Abf2p, and anti-Mgm101p antibodies. Mitochondria were lysed and mitochondrial nucleoids were enriched by sucrose gradient centrifugation according to Materials and Methods. A constant amount (50 μg) of the M fraction (M, lane 6), the top fraction from the sucrose gradient (TF, lane 7) and the 60/80% interface fraction containing mitochondrial nucleoids (NUC, lane 8) was analyzed as described above.
Figure 4
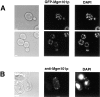
Mgm101p is localized to mitochondrial nucleoids in vivo. (A) W303 cells expressing mito-GFP-Mgm101p were grown in YPG, stained vitally with DAPI, and directly imaged as described in Materials and Methods. (B) W303 cells overexpressing Mgm101p were grown in YPG and were processed for indirect immunofluorescence with anti-Mgm101p and imaged as described in Materials and Methods. Bar, 2 μm.
Figure 5
Mgm101p binds DNA. Mitochondrial nucleoids were isolated as described in Materials and Methods. Nucleoid-associated proteins were dissociated and separated from mitochondrial DNA by incubation for 2 h in 1 M KCl followed by centrifugation where the salt extract was layered over a 30% sucrose cushion. The supernate containing high salt extracted proteins was dialyzed into Tris-EDTA buffer, pH 7.5, 200 mM NaCl (S, lane 1) and adsorbed to a native DNA cellulose column. Proteins bound to the column were eluted with 250 mM, 500 mM, and 1 M NaCl. The dialyzed high salt extract (S, lane 1), flow through (FT, lane 2), and fractions from 250 mM, 500 mM, and 1 M elutions (lanes 3–5) were analyzed by SDS-PAGE and immunoblotting.
Figure 6
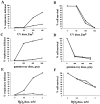
Mgm101p content in mgm101-2 cells decreases under nonpermissive conditions. A. Cultures of strain W303(pep4), JNY131(mgm101-2), and CS6-1D were grown in YPD. Total cell extracts were prepared by the rapid boiling method as described in Materials and Methods. 0.33 OD600 units of W303, pep4 (lane 1), JNY131 (lane 2) and CS6-1D (lane 3) was analyzed by SDS-PAGE and immunoblotted with anti-Mgm101p antibody. B. JNY131(mgm101-2) cells were grown to log phase in YPG and cells were harvested, split and washed into YPD medium at 24°C or YPD medium prewarmed at 37°C. These two cultures of JNY131(mgm101-2) were grown in YPD at 24°C and 37°C, respectively, and samples were collected every 2 h. Total cell extracts were prepared and analyzed as described in Part A.
Figure 6
Mgm101p content in mgm101-2 cells decreases under nonpermissive conditions. A. Cultures of strain W303(pep4), JNY131(mgm101-2), and CS6-1D were grown in YPD. Total cell extracts were prepared by the rapid boiling method as described in Materials and Methods. 0.33 OD600 units of W303, pep4 (lane 1), JNY131 (lane 2) and CS6-1D (lane 3) was analyzed by SDS-PAGE and immunoblotted with anti-Mgm101p antibody. B. JNY131(mgm101-2) cells were grown to log phase in YPG and cells were harvested, split and washed into YPD medium at 24°C or YPD medium prewarmed at 37°C. These two cultures of JNY131(mgm101-2) were grown in YPD at 24°C and 37°C, respectively, and samples were collected every 2 h. Total cell extracts were prepared and analyzed as described in Part A.
Figure 7
Mitochondrial nucleoid morphology and mtDNA replication are unaffected in mgm101-2 cells. W303TK and JNY131TK cells were cultured overnight at 25°C in YPG, washed and resuspended in YPD at 37°C. Aliquots of cells were taken at fixed time points after incubation at 37°C and either (A) fixed and labeled with DAPI or (B) labeled with BrdU. Representative samples at various time points are shown.
Figure 8
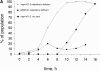
mgm101-2 cells are defective for the repair of oxidatively damaged mtDNA. (A and B) UV irradiation of W303 and JNY131(mgm101-2) cells was performed at indicated doses after cells were cultured in log phase at 37°C in YPD for 2 h and plated onto YPD media at 25°C. (C and D) Gamma ray treatment was performed at 4°C on cultures of W303 and JNY131(mgm101-2) cells that were grown overnight at 25°C in YPG, washed into saline solution and sonicated. (E and F) H2O2 treatment was performed at 25°C on cultures of W303 and JNY131 (mgm101-2) cells that were grown overnight in YPG, washed into YPD and treated with H2O2 at the indicated concentrations. Respiratory deficiency (A, C, and E) and cell survival (B, D, and F) were assessed as described in Materials and Methods. W303 cells are represented by circles and JNY131(mgm101-2) cells are represented by squares.
Similar articles
- The N-terminal intrinsically disordered domain of Mgm101p is localized to the mitochondrial nucleoid.
Hayward DC, Dosztányi Z, Clark-Walker GD. Hayward DC, et al. PLoS One. 2013;8(2):e56465. doi: 10.1371/journal.pone.0056465. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418572 Free PMC article. - A role for MHR1, a gene required for mitochondrial genetic recombination, in the repair of damage spontaneously introduced in yeast mtDNA.
Ling F, Morioka H, Ohtsuka E, Shibata T. Ling F, et al. Nucleic Acids Res. 2000 Dec 15;28(24):4956-63. doi: 10.1093/nar/28.24.4956. Nucleic Acids Res. 2000. PMID: 11121487 Free PMC article. - Mitochondrial DNA repair and damage tolerance.
Stein A, Sia EA. Stein A, et al. Front Biosci (Landmark Ed). 2017 Jan 1;22(5):920-943. doi: 10.2741/4525. Front Biosci (Landmark Ed). 2017. PMID: 27814655 Review. - Mitochondrial chromosome structure: an insight from analysis of complete yeast genomes.
Nosek J, Tomaska L, Bolotin-Fukuhara M, Miyakawa I. Nosek J, et al. FEMS Yeast Res. 2006 May;6(3):356-70. doi: 10.1111/j.1567-1364.2005.00016.x. FEMS Yeast Res. 2006. PMID: 16630276 Review.
Cited by
- The N-terminal intrinsically disordered domain of Mgm101p is localized to the mitochondrial nucleoid.
Hayward DC, Dosztányi Z, Clark-Walker GD. Hayward DC, et al. PLoS One. 2013;8(2):e56465. doi: 10.1371/journal.pone.0056465. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418572 Free PMC article. - Genetic instability in budding and fission yeast-sources and mechanisms.
Skoneczna A, Kaniak A, Skoneczny M. Skoneczna A, et al. FEMS Microbiol Rev. 2015 Nov;39(6):917-67. doi: 10.1093/femsre/fuv028. Epub 2015 Jun 24. FEMS Microbiol Rev. 2015. PMID: 26109598 Free PMC article. Review. - Yeast aconitase binds and provides metabolically coupled protection to mitochondrial DNA.
Chen XJ, Wang X, Butow RA. Chen XJ, et al. Proc Natl Acad Sci U S A. 2007 Aug 21;104(34):13738-43. doi: 10.1073/pnas.0703078104. Epub 2007 Aug 14. Proc Natl Acad Sci U S A. 2007. PMID: 17698960 Free PMC article. - Mitochondrial protein synthesis, import, and assembly.
Fox TD. Fox TD. Genetics. 2012 Dec;192(4):1203-34. doi: 10.1534/genetics.112.141267. Genetics. 2012. PMID: 23212899 Free PMC article. - Septin-containing barriers control the differential inheritance of cytoplasmic elements.
Tartakoff AM, Aylyarov I, Jaiswal P. Tartakoff AM, et al. Cell Rep. 2013 Jan 31;3(1):223-36. doi: 10.1016/j.celrep.2012.11.022. Epub 2012 Dec 27. Cell Rep. 2013. PMID: 23273916 Free PMC article.
References
- Alberts B, Herrick G. DNA-cellulose chromatography. Methods Enzymol. 1971;21:198–217.
- Birky CW. Transmission genetics of mitochondria and chloroplasts. Annu Rev Genet. 1978;12:471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials